PNL Volume
20
1988
RESEARCH
REPORTS
43
Table 1.
Segregation of triosephosphate isomerase phenotypes
|
Cross |
TPI phenotype* |
2 X
(1:2:1) |
||||
|
a |
ab |
b |
||||
|
JI 2018 x JI
1794
JI 1794 x slow
JI 1794 x
A783-161 |
9 19
11 |
19
32
26 |
9 12
16 |
0.02 1 .57
0.96 |
||
Designations: a = faster
migrating variant, ab = 3-banded phenotype, b = slower migrating
variant.
Table 2. Joint segregation of Le and
Tpi-p.
|
Cross |
N |
No. plants with designated
phenotype* |
x2 |
Recomb
Fract. |
|||||||
|
+ /aa |
+/ab |
+ /bb |
-/aa |
-/ab |
-/bb |
||||||
|
JI 1794 x slow JI 1794 x
161 |
63 53 |
19 11 |
32 23 |
0 5 |
0 0 |
0 3 |
12 11 |
63.0
21.7 |
1 +/- 1 15 +/-
5 |
||
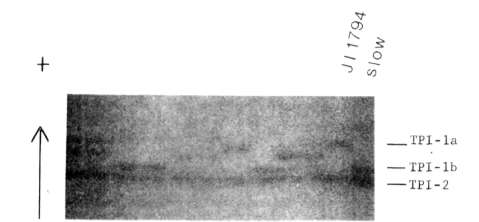
Fig. 1. Segregation of
triosephosphate isomerase phenotypes in the JI 1794 x slow progeny. The
parental phenotypes are shown on the left side of the gel. Anode is toward
top of figure,
*****