
Screening for field resistance to Powdery Mildew (Eyrsiphe polygoni D.C.) in the JI Pisum Collection
Ambrose, M.J. John Innes Centre, Norwich, UK
Powdery mildew is a serious disease in peas grown in areas with dry warm days and nights cool enough for dew formation. The infection causes a reduction in plant productivity, delays the drying out of plant tissues and can cause discoloration and off-flavors to the seed (1). Resistance was first reported by Harland in 1948 in material originating from Huancabamba in Northern Peru growing at an altitude of 1948m (2). This resistance (er1) has proved highly durable to date and Cousin reports in 1997 that the resistance had held up in France from when it was first introduced in1965 (3). Notes of resistant accessions in the John Innes Pisum collection made in the early 1980’s are still resistant today. The use of the resistance by breeders in the many regions of the world (including Australia, Canada and the US) where the disease is a problem has been steady since the 1970’s. In Europe this has accelerated as part of the drive for higher agronomic sustainability and implementation of changes in EU pesticide legislation (Council Directive 91/414 EEC) which is reducing the availability of fungicide treatments. Two further loci conferring resistance have been reported, er2 (4) and Er3 (5). While a number of research groups are focused on identifying, isolating and sequencing these loci, establishing the distribution of resistance in broader germplasm is still of interest in helping to target future screening for naturally occurring allelic variation.
Field resistant accessions in the JIC collection have been grown periodically as part of regeneration programs and opportunistically scored for mildew resistance/susceptibility. Resistance scores collected over a period of 26 years may include false positives where the resistant scores of some accessions may have been due to the lack of symptoms through disease avoidance such as early maturity. This study was undertaken to assess the current status of all accessions previously scored as showing field resistance in a single growing season and to explore the distribution and origins of that material.
Field resistance to powdery mildew (PMR) had previously been scored in 66 Pisum sativum accessions during the period 1982 to 2008. Five seeds of each accession were sown against wire with a susceptible control (JI 502: Rondo) sown at the start of the row and every 11th line position along the row. The trial was grown on experimental plots adjacent to the JIC. Peas have been continuously grown in this vicinity since the early 1970’s and infection each year is; therefore, effectively guaranteed.
Plants were scored at mid-podding at the start of June two weeks after mildew was first noted. This was well into the flowering and early podding period and white patches of mycelium were clearly visible on susceptible lines over the entire plant including pods. A second scoring was conducted in mid July when plants were senescing.
Resistant lines were cross-referenced to the output of a new structure analysis output based on a genotype dataset of 45 RBIP retro-element markers scored across the JI Pisum Collection (6). The recent availability of this output marks the start of a new era of more predictive management and selection of germplasm utilising the entire collection and contrast to earlier approaches where such detailed resolution was only feasible on subsets of lines which led to the development of core collections to represent the wider diversity.

Of the 66 lines, 60 lines were found to exhibit strong resistance with no visible spores over leaves, stipule, stems or pods. The second scores, conducted blind later in the growing season, confirmed the earlier scores. At this stage the resistant material was clearly visible as the only living green material in any of the pea trials. Six lines previously scored as resistant were found to be susceptible. It is not clear whether the resistance in these accessions had been overcome but the evidence of the remaining material suggests this is not the case. What is more likely is the fact that the previous resistant scores may have been false due to the material maturing before the infection could take hold or the infection being particularly light that year. A table of resistant accessions is presented in Table 1.
|
Combined order |
JI Acc. No. |
Name |
CTY |
Group |
|
NA |
1050 |
WIS-7104 |
USA |
NA |
|
NA |
2480 |
CGN 3352 |
PER |
NA |
|
NA |
1064 |
X 13463 |
PER |
NA |
|
26 |
2216 |
POWDERY MILDEW RESISTANT |
|
1.1 |
|
51 |
73 |
WBH 1238 |
|
1.1 |
|
268 |
1391 |
WBH 2138-wp |
USA |
1.2 |
|
411 |
2217 |
POWDERY MILDEW RESISTANT |
|
1.3 |
|
544 |
26 |
STIPULA-IMMINUATA-stim |
SWD |
1.4 |
|
634 |
229 |
P.SATIVUM-MALAYA |
UTK |
1.4 |
|
683 |
1749 |
OFI |
USA |
1.4 |
|
710 |
2302 |
B76-197 (STRATAGEM) |
FRN |
1.4 |
|
766 |
105 |
P.SATIVUM-AFGHANISTAN |
AFG |
1.5 |
|
835 |
2301 |
MEXIQUE 4 |
MEX |
1.5 |
|
897 |
1559 |
MEXIQUE 4 |
MEX |
1.5 |
|
947 |
1401 |
MANOA SUGAR |
|
1.6 |
|
1015 |
1180 |
MARX G-TYPE |
USA |
1.6 |
|
1068 |
1243 |
NEW LINE E.PERF./PMR |
USA |
1.7 |
|
1163 |
1239 |
DSP/PMR |
USA |
1.7 |
|
1327 |
1048 |
WIS-7102 |
USA |
1.7 |
|
1335 |
1069 |
WILTY-wil |
|
1.7 |
|
1501 |
1205 |
MISOG-2:tl |
USA |
2.1 |
|
1503 |
1203 |
MISOG-2:af |
USA |
2.1 |
|
1514 |
1207 |
MISOG-2:af,tl |
USA |
2.1 |
|
1542 |
1769 |
MELTON-af |
UTK |
2.1 |
|
1555 |
1568 |
CM-014F |
|
2.1 |
|
1560 |
1204 |
MISOG-2:st |
USA |
2.1 |
|
1575 |
1241 |
NEW ERA/PMR |
USA |
2.1 |
|
1610 |
1199 |
MISOG-1:af,tl |
USA |
2.1 |
|
1627 |
1567 |
EDISON |
|
2.1 |
|
1639 |
1201 |
MISOG-1:af,st,tl |
USA |
2.1 |
|
1698 |
1562 |
B277-457-1 |
|
2.1 |
|
1740 |
1237 |
SPRITE/PMR |
USA |
2.1 |
|
1745 |
1213 |
ERYLIS |
FRN |
2.1 |
|
1760 |
1424 |
VR74-1492-1 |
USA |
2.1 |
|
1766 |
2072 |
VIP |
|
2.1 |
|
1789 |
1194 |
MISOG-1:CONVENTIONAL |
USA |
2.1 |
|
1876 |
1049 |
WIS-7103 |
USA |
2.1 |
|
1910 |
1047 |
WIS-7101 |
USA |
2.1 |
|
1958 |
1766 |
BARTON-af,st |
UTK |
2.1 |
|
2011 |
1566 |
ALMOTA |
|
2.1 |
|
2017 |
1171 |
P.SATIVUM-INDIA |
IND |
2.1 |
|
2129 |
1412 |
MARLIN |
USA |
2.2 |
|
2174 |
1817 |
HJA 51902-af |
FIN |
2.2 |
|
2318 |
143 |
B268-394-3 |
|
2.2 |
|
2357 |
1780 |
TWIGGY-af |
USA |
2.2 |
|
2375 |
1128 |
P.SATIVUM-INDIA |
IND |
2.2 |
|
2443 |
1816 |
HJA 51893-af |
FIN |
2.2 |
|
2457 |
210 |
LUCKNOW BONIYA |
UTK |
2.2 |
|
2753 |
1214 |
TRIANON |
FRN |
3.1 |
|
2756 |
1951 |
P.SATIVUM-CHINA |
CHN |
3.1 |
|
2793 |
1557 |
P.SATIVUM-AFGHANISTAN |
AFG |
3.3 |
|
2799 |
92 |
P.SATIVUM-AFGHANISTAN |
AFG |
3.3 |
|
2802 |
96 |
P.SATIVUM-AFGHANISTAN |
AFG |
3.3 |
|
2809 |
101 |
P.SATIVUM-AFGHANISTAN |
AFG |
3.3 |
|
2812 |
95 |
P.SATIVUM-AFGHANISTAN |
AFG |
3.3 |
|
2813 |
102 |
P.SATIVUM-AFGHANISTAN |
AFG |
3.3 |
|
2814 |
100 |
P.SATIVUM-AFGHANISTAN |
AFG |
3.3 |
|
2856 |
1388 |
FENOMEN |
|
3.4 |
|
3027 |
2019 |
P.SATIVUM-LADAKH |
IND |
3.7 |
Three resistant accessions (JI 1050, JI 1064 and JI 2480) were not screened as part of the structure analysis and so could not be assigned to a group. This is unfortunate as JI 1050 and JI 1064 can both be traced back to the original resistant material from Peru first reported by Harland some 51 years ago (2,4). Resistant lines were found to be spread throughout the three major groups identified in the structure analysis (Fig. 1). The 12 accessions in group 1, which constitutes predominantly landraces, are relatively evenly spaced.
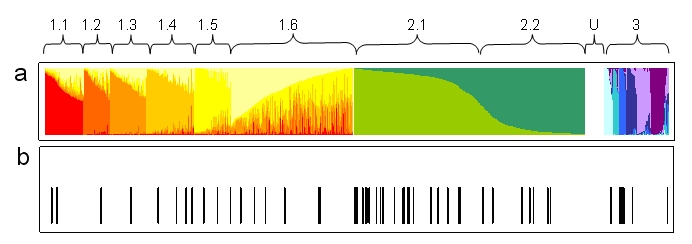
Figure 1. a. Output of structure analysis of 45 RPIB markers on 3029 accessions of the JIC Pisum Collection based on K=3,
b. Bars indicate the position of PMR accessions listed in Table 1.
The distribution of lines in group 2, which is predominantly composed of cultivars, is more concentrated in subgroup 2.1. This is largely due to the 7 near isogenic lines (MISOG1, MISOG2 series)(7), occurring in close order. The presence of resistant accessions in group 3 is of considerable interest. Group three has the strongest sub-group structure and all the accessions of wild species (P. fulvum, P. elatius) along with an array of P. sativum, including cultivated forms which show a higher degree of relatedness to these forms than the majority of P. sativum (6). Among this material are accessions from a range of countries including France, China and India and, most notably, a group of seven from Afghanistan. Collection site data are available for 8 of these from Afghanistan to China which are presented in Figure 2.
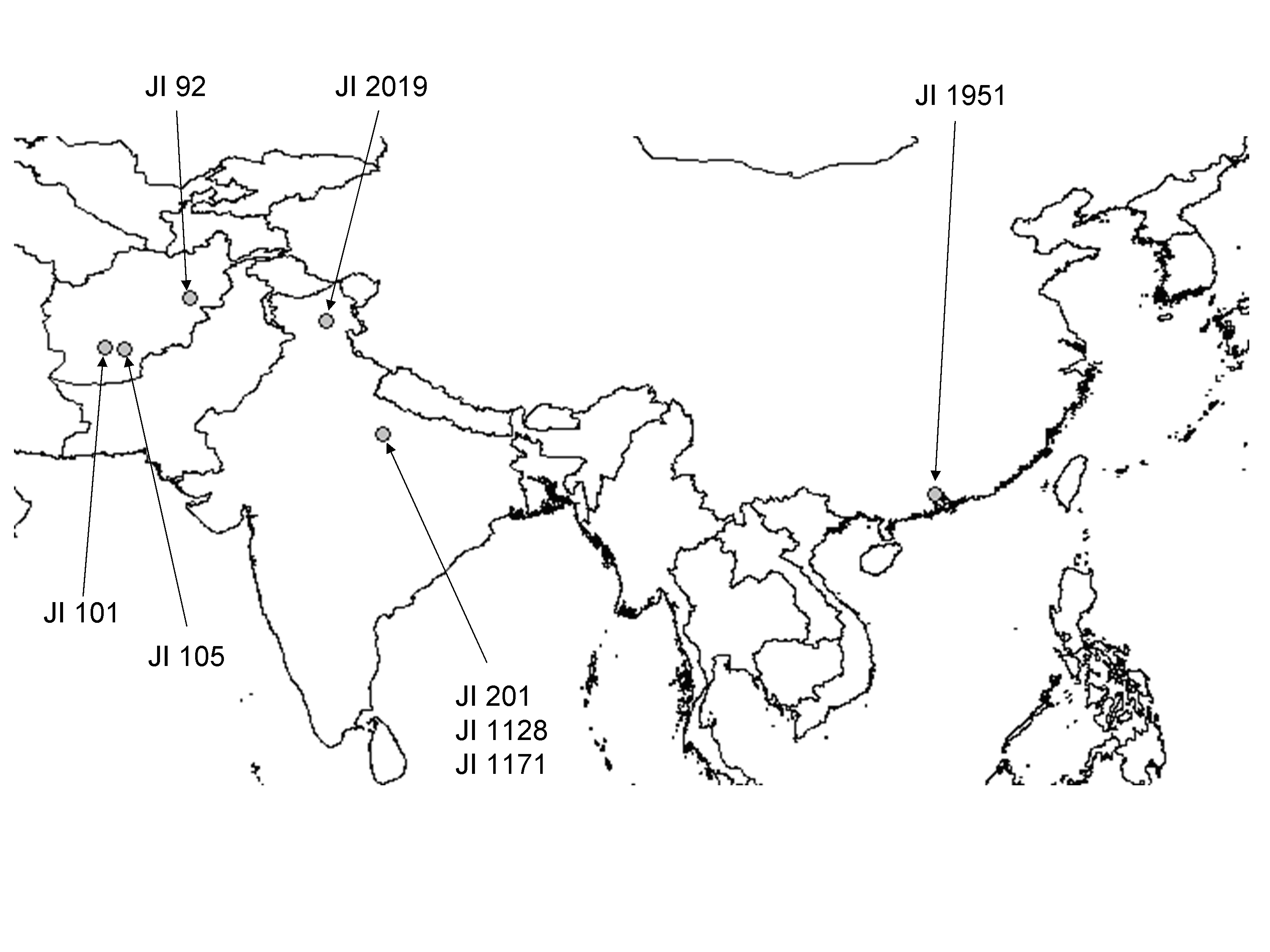
Figure 2. Collection sites of 8 accession showing field resistance to powdery mildew.
The geographic range over which these resistant lines from group 3 are spread is extremely large. This is the first such report of resistance to powdery mildew being reported in material from this region and raises some intriguing questions. The high altitude associated with the secondary region of diversity (Asiatic highland including the Hind Kusch) as the collection sites of two of these accessions (JI 92 and JI 2019 above 2000m) bears close parallels with the initial source of resistant germplasm in Peru. Both are associated with high altitude which would provide the warn days and cool nights favoured by the disease. There is no knowledge as to the genetic basis of the PMR observed in this material or how many resistance genes might be present in each accession. A detailed comparison will only become possible through lengthy genetic studies or comparisons of sequences from each of the resistance genes once they become known. Until that time it is impossible to establish whether the resistance found in the new world material was present in peas taken over to the Americas by early pioneers and is; therefore, derived from these old world sources outlined or whether the resistance is novel and emerged independently due to high selection pressure of the disease. Either way, the nature and genetic basis for the resistance reported for the first time here in this old world germplasm will clearly be of interest in future studies. All the germplasm cited above is available on request.
1. Hagedorn, D.J. 1984. Compendium of pea diseases, American Phytopathology Soc. p 20.
2. Harland, S. C. 1948. Heredity 2:263-269.
3. Cousin, R. 1997. Field Crops Research 53:111-130.
4. Heringa et al. 1968. Euphtyica, 18:163-169.
5. Fondevilla, S., Torres, A.M., Moreno, M.T. and Rubiales, D. 2007. Breeding Sci. 57:181-184.
4. Pierce, W.H. 1948. Phytopathology, 38:21.
5. Gritton, E.T. and Hagedorn, D.J. 1971. Crop Sci. 11:941.
6. Jing, R., Vershinin, A., Grzebyta, J., Shaw, P., Smýkal, P., Marshall, D., Ambrose, M.J., Ellis, T.H.N. and Flavell, A.J. 2009. BMC Evol. Biol.(submitted).
7. Marx, G.A. 1974. Pisum Newsl. 6:60.