Isolation of
Pseudo Response Regulator genes and evaluation as candidate genes
for photoperiod response loci
Liew, L.C.1, Hecht, V.1,
1University of
Weeden, N.2 and Weller, J.L.1
Introduction
The genetic
control of flowering in pea has been studied for more than five decades, and
several loci affecting photoperiod response are known (1). Mutations at the Sn, Dne, Ppd or Hr
loci result in early-flowering under short-day conditions (2, 3, 4, 5) whereas loss-of-function
mutations in the PhyA
or Late1 genes cause late flowering
under long-day conditions (6, 7). In arabidopsis,
many genes that affect photoperiodic flowering have a primary role in
regulation of the circadian clock, and we recently showed that Late1 is the pea ortholog
of the clock-related arabidopsis gene GIGANTEA (GI) (6). This study also showed that Late1 interacts genetically with Sn, and that sn mutant impairs
the diurnal expression rhythm of Late1
(6). This provides the first
direct evidence that Sn might be involved in
the clock mechanism in some way.
One potential
route to the identification of the Sn gene could be to assess homologs
of arabidopsis circadian clock-related genes as
candidates. We previously isolated several clock-related genes in pea (8), but
found that none of them mapped close to known photoperiod response loci.
However, the list of potential candidate genes for these loci has been extended
with recent identification of additional clock-related genes in arabidopsis. Isolation of the corresponding pea genes has
been greatly assisted by recent progress in sequencing of the medicago genome and advances in comparative genome analysis
between medicago and pea (9, 10, 11).
Several recent
studies have examined the contribution of the PSEUDO RESPONSE REGULATOR (PRR)
gene family to the circadian clock mechanism (12, 13, 14, 15). This family includes the core
clock gene TIMING OF CAB EXPRESSION 1
(TOC1) and four other members; PRR9, PRR7, PRR5,
and PRR3. All five genes show diurnal and circadian regulation,
with distinct peaks of expression that occur sequentially every 2 hours after
dawn (12). This finding has suggested
that like TOC1, other members of the “PRR
quintet” might also form part of the central oscillator. We recently observed
that the effect of the sn
mutant on the expression of LATE1 is
similar to the effect of the prr5 prr7 prr9 triple mutant on GI in arabidopsis
(16), raising the possibility that
in some respects Sn
might act similarly to PRR genes. We
therefore set out to isolate PRR
genes in pea, in order to examine their potential role within the pea circadian
clock, and also their potential identity as candidate genes for photoperiod
response loci.
Materials
and Methods
Sequences of PRR
homologs from Medicago truncatula and other species were obtained using tBLASTn searches of the Genbank
database (http://www.ncbi.nlm.gov) and the medicago
EST database at http://compbio.dfci.harvard.edu. To isolate members of the PRR
gene family in pea, degenerate primers were designed within conserved domains
using the CODEHOP strategy (http://blocks.fhcrc.org/codehop.html) (17). The full length PsPRR37
and partial PsPRR59 cDNA were obtained by 5’
and 3’ RACE-PCR using the BD-SMART RACE cDNA
amplification kit (CLONTECH). Protein alignments of various PRRs
were performed with ClustalX (18) and adjusted using GENEDOC
(Nicholas et al. 1997; http://www.psc.edu/biomed/genedoc). Relationships among PRR
amino acid sequences were determined using phylogenetic
analyses in PAUP* 4.0b10 (http://paup.csit.fsu.edu).
The origin of the
WT line NGB5839 (cv. Torsdag le-3) and the sn-2 and sn-4 mutants have been described
previously (19, 6). The sn-3 mutant is an additional recessive mutant isolated in the same
screen as sn-4 (6). All plants were grown in the
Hobart phytotron, using previously-described growth
media, light sources and phytotron conditions (6).
Information and
approximate map positions of pea genes in the bottom half of LGVII was obtained
from several published maps (20, 21, 22). To identify medicago homologs of pea genes in
this region, tBLASTx searches were performed against
the medicago genomic database at the J. Craig Venter
Institute (http://www.jcvi.org/cgi-bin/medicago/index.cgi). The map positions of
relevant genes were obtained by using Medicago Genome
Browser (http://gbrowse.jcvi.org/cgi-bin/gbrowse/medicago_imgag/).
Results
and Discussion
Identification of PRR homologs in Medicago truncatula
PRR proteins are
characterized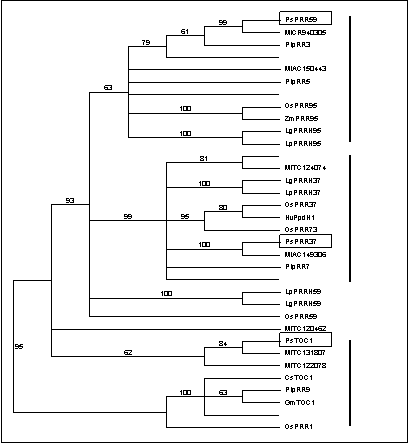 by two conserved domains; the Pseudo-regulator
(PR) domain and the CONSTANS, CONSTANS-LIKE, and TOC1 (CCT) domain (12). Several reports have
identified PRR genes in a number of species including rice (23), Lemna
gibba, L. paucicostata
(24) and Populus
trichocarpa (25). Phylogenetic
analysis shows that these genes fall into three major groups, which can be
designated as PRR1, PRR59 and PRR37, on the basis of the arabidopsis sequences they include (Figure 1). Two
accessions from Lemna and one from rice (OsPRR59)
show affinity to the PRR59 clade at the sequence level, but fall outside this group in
the phylogenetic analysis, most probably because they
do not contain complete sequences for both conserved domains.
by two conserved domains; the Pseudo-regulator
(PR) domain and the CONSTANS, CONSTANS-LIKE, and TOC1 (CCT) domain (12). Several reports have
identified PRR genes in a number of species including rice (23), Lemna
gibba, L. paucicostata
(24) and Populus
trichocarpa (25). Phylogenetic
analysis shows that these genes fall into three major groups, which can be
designated as PRR1, PRR59 and PRR37, on the basis of the arabidopsis sequences they include (Figure 1). Two
accessions from Lemna and one from rice (OsPRR59)
show affinity to the PRR59 clade at the sequence level, but fall outside this group in
the phylogenetic analysis, most probably because they
do not contain complete sequences for both conserved domains.
 BLAST
searches of the Medicago truncatula
genomic (NCBI) and EST databases identified seven distinct PRR sequences, including three genomic and four ESTs.
Additional EST contigs corresponding to three of the
genomic sequences were also identified. Four of these sequences were predicted
to encode full-length PRR proteins, including two in the PRR59 clade and two in the PRR37 clade.
The remaining three EST sequences were only partial. Two distinct ESTs from the 5’ region grouped with the partial pea TOC1 sequence described previously,
while the other from the 3’ region did not show a clear relationship to any
other PRR.
BLAST
searches of the Medicago truncatula
genomic (NCBI) and EST databases identified seven distinct PRR sequences, including three genomic and four ESTs.
Additional EST contigs corresponding to three of the
genomic sequences were also identified. Four of these sequences were predicted
to encode full-length PRR proteins, including two in the PRR59 clade and two in the PRR37 clade.
The remaining three EST sequences were only partial. Two distinct ESTs from the 5’ region grouped with the partial pea TOC1 sequence described previously,
while the other from the 3’ region did not show a clear relationship to any
other PRR.
The three medicago BAC contigs containing
the genomic PRR sequences have all
been assigned map positions (www.medicago.org), on chromosome 3 (CR940305),
7 (AC150443) and 4 (AC149306). These positions predict positions for the
corresponding pea sequences in the middle of LGIII, near the top of LGV, and in
the bottom half of LGVII, respectively. We noted in particular that AC149306
was located in a region of chromosome 4 corresponding to the region of pea
LGVII known to contain the Sn locus. Sn was previously
reported to show close linkage with the amylase locus Amy1 (26) and was mapped between isozyme loci Aldo and Gal2 (27) on the lower section of pea
linkage group VII. The similar positions of MtPRR59
(CR940305) and the Dne
locus also suggested a possible candidate gene relationship.
Isolation of PRR homologs from pea
Using degenerate
primers targeting the PR and CCT domains two distinct fragments from the PR
domain were amplified by PCR. These sequences were extended to both 5’ and 3’
ends using RACE PCR to obtain one partial (70%) and one full-length coding
sequence. Phylogenetic analysis showed that these
sequences belonged the PRR59 and PRR37 clades, respectively, and they were designated as PsPRR59 and PsPRR37. Figure 1 shows that these sequences are apparent orthologs of the medicago PRR genes on CR940305 and AC149306.
Mapping of PRR genes and
evaluation of PsPRR37 as a candidate for Sn
Sequencing of PRR59 and PRR37 from mapping parents JI1794 and “Slow” identified
polymorphisms that were used to map both genes in the RIL population derived
from these parents (22). The results confirmed
positions for PRR59 in LGIII near Dne and for PRR37 in LGVII, consistent with the
positions predicted by the location of the orthologous
medicago genes. The relationship between PRR59 and Dne was not explored further, as Dne has been
identified as the pea ortholog of the arabidopsis ELF4
gene (28). However, for PRR37, no recombination was detected with
the Amy locus on LGVII, and as Amy
was previously noted to be tightly linked to Sn (26), this indicated that PRR37 is in the region of Sn.
In order to carry
out fine mapping of Sn,
we also generated a new mapping population derived from a cross between the sn-4 mutant in the NGB5839 background
and cv, Térèse.
Unfortunately, we found that the coding region of PRR37 was identical in NGB5839 and Térèse,
precluding the straight-forward mapping of PRR37
relative to Sn
in this cross. Instead, the entire coding sequence of PRR was determined from the three known induced sn mutants
and from their isogenic wild-type lines Borek (sn-2) and
NGB5839 (sn-3 and sn-4). In all three cases the PRR37 coding sequence was identical in
mutant and wild-type, indicating that the sn phenotype does not
result from a mutation that affects PRR37
protein structure. It will obviously be of interest in future to isolate
flanking sequence of PRR37 and
identify an appropriate polymorphism that will allow the cosegregation
of PRR37 and Sn to be directly tested.
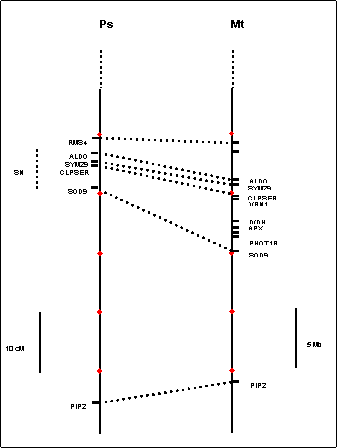
A comparative map of the Sn
region
As an aid to
future mapping studies in the region of Sn, we generated a comparative map using markers anchored in
published pea linkage maps and the medicago physical
map (Figure 2). This identified a broad region likely to contain Sn, bounded approximately by the Aldo and Sod9 genes. In medicago, the physical map
of this region is estimated to span approximately 5Mb, although it still
contains several gaps where adjacent BAC contigs are
not yet joined. We are now using this information as a basis for the design of
additional markers for the mapping of Sn and for the identification of other potential candidate
genes. The first step will be the mapping of Sn relative to other markers shown.
1. Weller, J. L. 2007.
Pisum Genetics 39:1-8.
2. King, W. M. and Murfet, I. C. 1985.
Annals of Botany 56:835-846.
3. Murfet, I. C. 1971. Heredity 27:93-110.
4. Murfet, I. C. 1973. Heredity 31
(2):157-164.
5. Murfet, I. C. and Taylor, S. A. 1999.
The Journal of Heredity 90 (5):548-550.
6. Hecht, V., Knowles, C. L., Vander Schoor,
J. K., Liew, L. C., Jones, S. E., Lambert, M. J. M.
and Weller, J.L. 2007. Plant Physiology 144 (2):648-661.
7. Weller, J. L., Batge, S. L., Smith, J.
J., Kerckhoffs, L. H. J., Sineshchekov,
V. A., Murfet, I. C. and Reid, J.B. 2004. Plant
Physiology 135 (4):2186-2195.
8. Hecht, V., Foucher, F., Ferrandiz, C., Macknight, R.,
Navarro, C., Morin, J., Vardy, M. E., Ellis, N., Beltran, J. P., Rameau, C. and
Weller, J. L. 2005. Plant Physiology 137 (4):1420-1434.
9. Choi, H. K., Mun,
J. H., Kim, D. J., Zhu, H., Baek, J. M., Mudge, J., Rob, J., Ellis, N., Doyle, J. and Kiss, G. B.
2004. Proceedings of the National Academy of Sciences of the United States of
America 101:15289-15294.
10. Kalo, P., Seres,
A., Taylor, S. A., Jakab, J., Kevei,
Z., Kereszt, A., Endre, G.,
Ellis, T. H. N. and Kiss, G.B. 2004. Molecular Genetics and Genomics
272:235-246.
11. Zhu, H., Choi, H.-K., Cook, D. R. and
Shoemaker, R. C. 2005. Plant Physiology 137 (4):1189-1196.
12. Matsushika, A., Makino, S., Kojima, M.
and Mizuno, T. 2000. Plant and Cell Physiology 41 (9):1002-1012.
13. Matsushika, A., Murakami, M., Ito, S., Nakamichi, N., Yamashino, T. and
Mizuno, T. 2007. Bioscience, Biotechnology and Biochemistry 71 (2):527-534.
14. Nakamichi, N., Kita, M., Niinuma, K., Ito, S., Yamashino,
T., Mizoguchi, T. and Mizuno, T. 2007. Plant and Cell
Physiology 48:822-832.
15. Salome, P. A. and McClung, C. R. 2005. The Plant Cell 17
(3):791-803.
16. Nakamichi, N., Kita, M., Ito, S., Yamashino, T. and Mizuno, T. 2005. Plant and Cell
Physiology 46 (5):686-698.
17. Rose, T. M., Schultz, E. R., Henikoff, J.
G., Pietrokovski, S., McCallum, C. M. and Henikoff, S. 1998. Nucleic Acids Research 26 (7):1637-1644.
18. Thompson, J. D., Gibson, T. J., Plewniak,
F., Jeanmougin, F. and Higgins, D. G. 1997. Nucleic
Acids Research 24:4876-4882.
19. Arumingtyas, E. L. and Murfet, I. C. 1994. Journal of Heredity 85:12-17.
20. Aubert, G., Morin, J., Jacquin, F., Loridon, K., Quillet, M. C., Petit, A., Rameau, C., Lejeune-He´naut,
I., Huguet, T. and Burstin,
J. 2006. Theoretical and Applied Genetics 112:1024-1041.
21. Laucou, V., Haurogne., K., Ellis, N. and
Rameau, C. 1998. Theoretical and Applied Genetics 97:905-915.
22. Weeden, N. F., Ellis, T. H. N.,
Timmerman-Vaughan, G. M., Swiecicki, W. K., Rozov, S.
M. and Berdnikov, V. A. 1998. Pisum Genetics 30:1-4.
23. Murakami, M., Ashikari, M., Miura, K., Yamashino, T. and Mizuno, T. 2003. Plant and Cell
Physiology 44 (11):1229-1236.
24. Miwa, K., Serikawa, M., Suzuki, S.,
Kondo, T. and Oyama, T. 2006. Plant and Cell Physiology
47 (5):601-612.
25. Tuskan, G. A., DiFazio,
S., Jansson, S., Bohlmann,
J., Grigoriev, I., Hellsten,
U., Putnam, N., Ralph, S., Rombauts, S., Salamov, A., Schein, J., Sterck,
L., Aerts, A., Bhalerao, R.
R., Bhalerao, R. P., Blaudez,
D., Boerjan, W., Brun, A.,
Brunner, A., Busov, V., Campbell, M., Carlson, J., Chalot, M., Chapman, J., Chen, G. L., Cooper, D., Coutinho, P. M., Couturier, J., Covert, S., Cronk, Q., Cunningham, R., Davis, J., Degroeve,
S., Dejardin, A., dePamphilis,
C., Detter, J., Dirks, B., Dubchak,
I., Duplessis, S., Ehlting,
J., Ellis, B., Gendler, K., Goodstein, D., Gribskov, M., Grimwood, J., Groover, A., Gunter, L., Hamberger,
B., Heinze, B., Helariutta,
Y., Henrissat, B., Holligan,
D., Holt, R., Huang, W., Islam-Faridi, N., Jones, S.,
Jones-Rhoades, M., Jorgensen, R., Joshi, C., Kangasjarvi,
J., Karlsson, J., Kelleher, C., Kirkpatrick, R., Kirst, M., Kohler, A., Kalluri,
U., Larimer, F., Leebens-Mack, J., Leple, J. C., Locascio, P., Lou,
Y., Lucas, S., Martin, F., Montanini, B., Napoli, C.,
Nelson, D. R., Nelson, C., Nieminen, K., Nilsson, O.,
Pereda, V., Peter, G., Philippe, R., Pilate, G., Poliakov, A., Razumovskaya, J.,
Richardson, P., Rinaldi, C., Ritland,
K., Rouze, P., Ryaboy, D., Schmutz, J., Schrader, J., Segerman,
B., Shin, H., Siddiqui, A., Sterky,
F., Terry, A., Tsai, C. J., Uberbacher, E., Unneberg, P., Vahala, J., Wall,
K., Wessler, S., Yang, G., Yin, T., Douglas, C., Marra, M., Sandberg, G., Van de Peer, Y. and Rokhsar, D. 2006. Science 313 (5793):1596-1604.
26. Weeden, N. F., Kneen,
B. E. and Murfet, I. C. 1988. Pisum Newsletter 20:49-51.
27. Murfet, I. C. and Sherriff, L. J. 1996. Pisum Genetics 28:13-14.
28. Liew, L. C., Hecht, V., Laurie, R. E.,
Knowles, C. L., Vander Schoor, J. K., Macknight, R. C. and Weller, J. L. 2009. Plant Cell:tpc.109.067223.