Pisum Genetics
2006—Volume 38
Research Papers
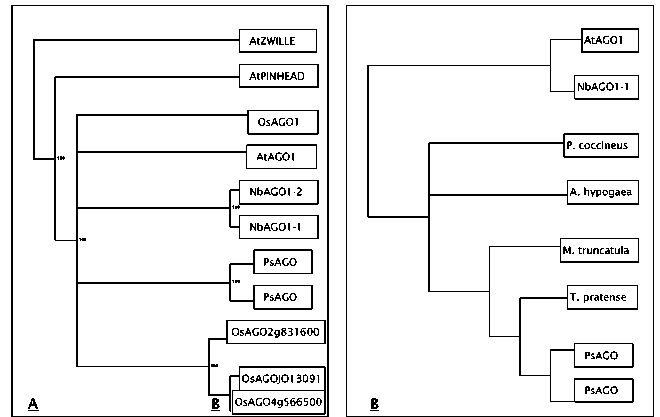
Fig. 1. Phylogenetic relationships among the Argonaute sequences based on PAUP analysis. A) The strict consensus tree for the full-length plant Argonaute genes available in the literature and B) the strict consensus tree for the comparison of approximately 600 bp of coding sequence at the 3’ end of an AGO gene from each of the legume species listed. The Arabidopsis and Nicotiana sequences were used as outgroups for the latter analysis.
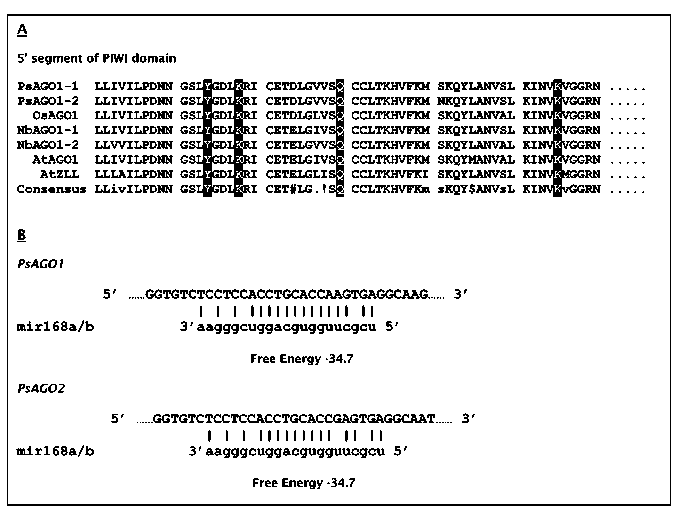
and PINHEAD sequences. The Pisum sequences were most closely related to each other, with the AtAGO1 sequence being the next most similar, confirming the results from the protein sequence analysis. Although two AGO1 genes are also found in Nicotiana benthamiana, the presumed duplication producing those two sequences apparently occurred independently of the duplication forming the two Pisum sequences. PAUP analysis of the partial coding sequences available from other legume species produced a tree that mirrored the established phylogeny of the taxa being compared (Fig. 1B). Two most parsimonious trees were detected. Only the strict consensus tree is shown in Fig. 1B. These results support the hypothesis that only AGO1 members of the Argonaute gene family are being compared in this analysis.
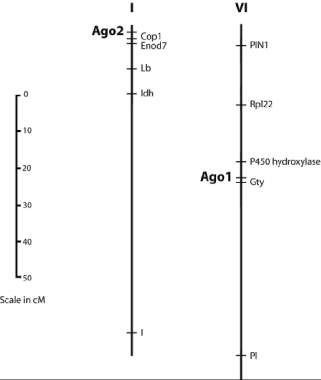
The PsAGO1 primers amplified a product of approximately 1500 bp that displayed polymorphism between JI 1794 and the domesticated lines when cut with the restriction enzyme HaeIII . The position of the locus Ago1 was determined to be on linkage group (LG) VI about 4 cM from Gty (Table 2). In the JI 1794 x ‘Slow’ linkage map, the region around Gty appeared to be compressed, with 10 or more markers within 5 cM of Gty. The order of the markers was ambiguous, and it could not be determined on which side of Gty Ago1 was located. The data from the second RI population indicated Ago1 lay distal to Gty, towards Rpl22 (Fig. 2).
The PsAGO2 primers amplified a fragment of about 2000 bp in the domesticated lines but did not amplify a product in JI 1794. This presence/absence polymorphism could be easily scored without need for CAPS analysis. Ago2 was found to be the most distal marker on the upper arm of LGI in both RI populations (Fig 2). The combined data from both populations placed Ago2 approximately 3 cM from Cop1 (Table 2).
5