Pisum Genetics
2006—Volume 38
Research Papers
A gene for stem fasciation is localized on linkage group III
Sinjushin, A.A., Konovalov, F.A. and Gostimskii, S A.
Fasciation is one of the most widespread abnormalities of higher plant development. An understanding of the inheritance of the trait is very important, not only for theoretical purposes dealing with genetic control of meristem activity but also for practical use. Stem and fruit fasciation is used as an agriculturally valuable trait in selection of many species including pea (Pisum sativum L).
Genetics Dept. Biolog. Faculty of Moscow State Univ., Moscow, Russia

Fig. 1. Fasciated plant of “Shtambovy” mutant line (a) and scanning electronic microphotographs depicting stem apical meristems of “Shtambovy” mutant (b) and wild type plant (Nemchinovsky cultivar, c). Scale bar =
100µ.
The peculiarities of genetic control of fasciation in pea are still being discussed. There are few genes responsible for fasciation development; these genes form the
Table 1. Analysis of segregation at single loci in an F
2
population. A – homozygote as the first parental line (‘Shtambovy’), B – homozygote as the second parental line (WL1238), H – heterozygote, N – total number of plants analyzed.
fasciata family although little is known about their structure, protein products and even localization on the genetic map. The gene Fa (or Fa1 as was proposed by Swi§cicki and Gawlowska (8)) is localized in linkage group IV (4), Fa2 is in LG V (8) and Fas is supposed to be associated with LG III (1).
The fasciated mutant ‘Shtambovy’ was produced by induced chemical mutagenesis
|
Loci
|
A
|
|
H
|
B
|
N
|
X2 (P>0.05)
|
||
|
Egl1
|
27
|
|
58
|
29
|
114
|
0.11
|
||
|
PK4
|
25
|
|
32
|
25
|
82
|
3.95
|
||
|
Pepcn
|
18
|
|
35
|
23
|
76
|
1.13
|
||
|
Le
|
|
91
|
|
27
|
118
|
0.28
|
||
|
Fas
|
|
90
|
|
30
|
120
|
0.00
|
||
(ethylmethane sulfonate) from the cultivar ‘Nemchinovsky’ (6). This mutant exhibits strong
features of fasciation such as stem flattening, phyllotaxis abnormalities, clustering of axillary racemes on top of the stem, etc. (Fig. 1a). Such phenotype is connected with stem apical meristem enlargement which can be seen with usage of scanning electron microscopy. The apex of mutants becomes ridge-like (Fig. 1b) instead of hemispheric in wild-type plants (Fig. 1c) thus producing ribbon-like stem with multiple bundles and a striated surface. The morphology, anatomy and growth characteristics of fasciated plants compared with normal ones have been previously described (7).
The fasciation in a new mutant line is caused by a recessive mutation in a single gene (see Table 1).
Allelism tests revealed that the gene responsible for fasciation in ‘Shtambovy’ is not allelic to gene Fa from JI 5 (‘Mummy Pea’): all F1 plants from cross ‘Shtambovy’ x JI 5 were non-fasciated.
In order to determine the possible relationship between ‘Shtambovy’ mutation and
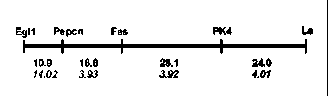
genes Fas and Fa2, an effort was made to localize the new fasciata locus on the pea linkage map. The F1 and F2 progeny of a cross ‘Shtambovy’ x WL
Fig. 2. Region of LG III containing gene Fas. Top numbers are genetic distances (cM), bottom numbers (in italics) are meanings of LOD score.
19