Pisum Genetics
2006—Volume 38
Research Papers
A mitochondrial DNA marker frequently found in wild peas
Bogdanova, V.S., and Kosterin, O.E. Inst. of Cytol. and Genet. of the Siberian Dept.
Russian Acad. of Sci., Novosibirsk, Russia
Earlier we reported on a chloroplast DNA marker polymorphic in wild peas (1). In the present work a mitochondrial DNA marker is characterized in a set of accessions which includes 31 wild and 18 cultivated samples (see Appendix). The list of accessions tested is extended compared to that reported in (1). The primers were designed 5'-TGGTAATTGGTCTGTTCCGATTCT-3' and 5'-
CCACTGCTTGAAGTGATTGTTACG-3' to match the nucleotide sequence of the EMBL X14409 accession (pea mitochondrial coxI gene for cytochrome oxidase subunit I). A fragment of 1200 bp was amplified using the following cycling parameters: initial denaturation at 95 C for 1 min followed by 38 cycles including denaturation at 94oC 59 sec, annealing at 56oC 45 sec, elongation at 72oC 1 min. Five microliters of the reaction were treated with one 1 unit of PsiI endonuclease for 2 hours at 37oC in a volume of 12 |ol and electrophoresed in 1.5% agarose gel in TAE buffer. We found a polymorphism for the presence/absence of the
PsiI recognition site, so that PCR products obtained from some accessions were digested into two fragments of 260 and 940 bp, while other samples produced a single band of 1200 bp.
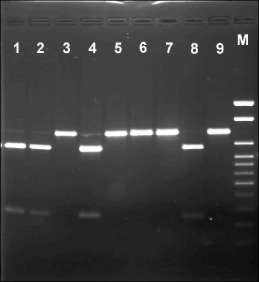
We assayed 49 specimens of wild and cultivated peas, which represent most of the presently recognised taxa. The digestion profiles of PCR products amplified from the cox1 gene of some pea accessions are shown in Fig. 1.
We found that the restriction site for PsiI in the mitochondrial gene cox1 is present in all eight accessions of Pisum fulvum Sibth et Smith and in all four accessions of P. sativum ssp. abyssinicum. Following Townsend (2) and Davis (3), we consider all wild representatives of Pisum sativum L. (except for P. sativum ssp. abyssinicum (A. Br.) Berger) within the same subspecies P. sativum ssp. elatius (Bieb.) Schmahl. sensu lato. In total we analysed 23 accessions of this subspecies and found that 11 of them have the restriction site
while 12 do not (see Appendix). The restriction site was found in neither accession of P. s. ssp. transcaucasicum Govorov examined nor in any of 12 accessions of P. s. ssp. sativum. Noteworthy, in 44 of 49 accessions analyzed, the presence of the recognition site for PsiI in the mitochondrial coxI gene coincided with the presence of the recognition site for AspLEI endonuclease in the plastid gene rbcL (1, and unpublished), the studied fragments being amplified from the same samples of genomic DNA extracted from a single plant for each accession. The five exceptions were VIR320*,
Fig. 1. Restriction fragments formed after PsiI digestion of PCR-amplified cox1 gene from some pea samples. Lanes: 1—VIR2759, P.s. ssp. abyssinicum; 2—WL1446, P.s. ssp. abyssinicum; 3—VIR4871, P.s. ssp. transcaucasicum; 4—WT6, P.s. ssp. abyssinicum; 5—VIR3954, P.s. ssp. sativum; 6—VIR2593, P.s. ssp. sativum; 7—VIR3429, P.s. ssp. sativum (“Pisum jomardi”?); 8—
VIR320*, P.s. ssp. elatius; 9—VIR1975, P.s. ssp. sativum; M—molecular weight marker 100-1000 bp + 1.5 kb + 2 kb.
VIR2123, L90, Ps008-120689-0202 (which had the restriction site in the mitochondrial coxI but not the restriction site in the plastid rbcL), and accession VIR1975 in which the situation was opposite. We conclude that the restriction site in the coxI gene, as well as that in the plastic gene rbcL, were both present in the closest common ancestor of the genus Pisum. Both have been lost in at least one lineage of P. sativum ssp. elatius, and this lineage gave rise to the cultivated P. sativum ssp. sativum.
1