Fine
mapping of the gene Crt of Pisum
sativum L.
controlling root morphogenesis and being involved
in interactions with symbiotic microorganisms
Kuznetsova, E.V.1, Tzyganov, V.E.1, Pinaev, A.G.1, 1All-Russia Res. Inst. for Agric. Microbiol. RAAS
Borisov A.Y.1 and Tikhonovich, I.A.1
Moffet, M.D.2 2Dept. of Plant Sci. and Plant Path.
Montana State Univ.,
The pea mutation curly roots was produced by chemical mutagenesis of the laboratory line SGE. The mutation was shown to be a recessive allele at a locus designated Crt (Tsyganov et al., 2000). Homozygous recessive plants show alterations in root morphology. These plants form a very compact root system with strongly curled roots in a substrate of high density (Tsyganov et al., 2000), decreased number of symbiotic root nodules and increased rate of formation of arbuscular mycorrhiza (in comparison with initial line SGE) (Alexander Zhernakov, personal communication). Cloning and sequencing Crt would permit us to better understand the function of this gene through being able to predict its molecular product. As an initial step for positional cloning, accurate and detailed mapping of the gene was performed. We were particularly interested in using molecular markers based on the primary sequences of expressed genes to exploit synteny between Pisum sativum L. and Medicago truncatula Gaertn.
The locus Crt was previously placed on pea linkage group V (Tsyganov et al., 2000).
To determine the map position of Crt accurately, molecular markers based on primary sequences of the genes Paal2 and Enol, as well as RAPD marker P603B, were developed. Specific primers for amplification of the part of phenylalanine ammonia lyase 2 (Paal2) gene (designed within the frames of joint COBASE project) and the primer for creation RAPD marker P603B were kindly provided by Dr. N. Weeden, Montana State University, USA. Primary sequence analysis of the amplified part of the gene Paal2 indicated restriction site polymorphism (for endonuclease Hae III) between lines NGB1238, RT9 and SGEcrt, allowing the creation of a CAPS marker. Moreover, line RT9 carryied a 47 bp deletion in the exon region of the gene Paal2 and differences in PCR product size could be easily detected in 1% agarose gel electrophoresis. To our knowledge primary sequence of the enolase gene of pea has yet to be determined. Enol primers creation was based on the comparison of primary sequences of Enolase genes of Medicago truncatula, Glycine max, Lupinus luteus, Ricinus communis and Arabidopsis thaliana. Amplified products of two lines, SGEcrt and NGB1238, were sequenced and absence of Hpa II restriction site in the amplified part of the enolase gene of the line SGEcrt was exploited. RAPD marker P603B is 20 bp length and provides more specific amplification and thus an opportunity to use the marker on other pea genotypes.
For Paal2 and Enol standard PCR protocol in 20-ml reaction mix was performed (94oC-4 min; 35 of 94oC- 30 sec, 60o C and 58oC, respectively, - 45 sec, 72oC- 1 min; 72oC 25 min; 4oC hold). Paal2 primer sequences: forward- 5’CAATAACATCAAAGTGAGTGACT, reverse- 5’GCTGAAGTTATGCAAGGGAAACC; Enol primer sequences: forward- 5’AGGATGACTGGGAGCACTATG, reverse- 5’CCAAGCTCCTCCTCAATTC. PCR protocol for P603B: 94oC- 4 min; 35 of 94oC- 30 sec, 37oC – 1 min, 72oC- 1 min; 72oC -7 min; 4oC hold. P603B primer sequence: 5’TGGAGTATATTCGAAGCTCG.
In addition to Paal, Enol and P603B, two morphological markers r and tl positioned near the locus of interest were used. Polymorphism for all markers was detected between the mutant line and laboratory line NGB1238. An F2 population of 103 individuals derived from a cross between the mutant line and NGB1238 was scored for all markers listed above. Plants homozygous for the crt mutant were identified by growing all plants on quartz sand. Two leaves from each plant were collected for DNA isolation. Linkage calculations were done using S. M. Rozov’s program CROSS.
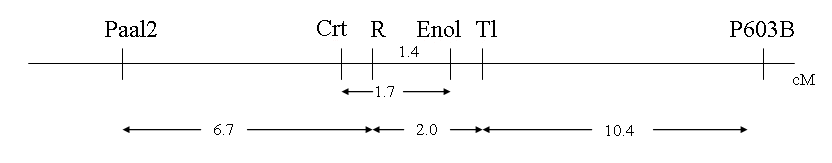
Table 1 presents the joint segregation analysis for most pairwise comparisons. Genes Crt and r were in the repulsion phase and the results obtained possessed a low P value for deviation from random assortment. The linkage data suggested the order of markers presented in Fig. 1, with Paal-2 at one boarder of the approximately 20 cM region illustrated and P603B at the other.

Our results position the locus Crt with more precision, an important step for further high-resolution mapping and map-based cloning of the gene of interest with the use of synteny between pea and Medicago truncatula Gaertn.
Acknowledgment: This research was financially supported by Russian Academy of Agricultural Sciences, grants of RFBR (04-04-48462), President of Russia (HIII-1103.2003.04, MD 350-2003.04), CRDF (ST-012-0) and COBASE program of NSF USA
1. Tsyganov, V.E., Pavlova, Z.B., Kravchenko, L.V., Rozov, S.M., Borisov, A.Y. and Lutova L.A., Tikhonovich I.A. 2000. Ann. Bot. 86: 975-981.
|
Table 1. Segregation data in F2 progeny of the cross lines SGEcrt ´ NGB 1238 |
||||||||||
|
Gene pair |
Number of progeny |
Linkage, cM |
Joint c2 |
P(0.5) |
||||||
|
AB |
AHz |
Ab |
aB |
aHz |
ab |
Total |
||||
|
r - tl |
78 |
- |
2 |
0 |
- |
23 |
103 |
2.01 ± 1.40 |
92.39 |
P<0.0001 |
|
P603B - r |
47 |
- |
1 |
7 |
- |
14 |
61 |
12.14 ± 4.24 |
35.82 |
P<0.0001 |
|
P603B - tl |
47 |
- |
1 |
6 |
- |
15 |
61 |
10.41 ± 3.92 |
39.44 |
P<0.0001 |
|
Crt - Enol |
18 |
37 |
1 |
0 |
0 |
10 |
66 |
1.73 ± 1.61 |
58.93 |
P<0.0001 |
|
Enol - r |
11 |
36 |
1 |
0 |
0 |
17 |
65 |
1.41 ± 1.47 |
60.11 |
P<0.0001 |
|
Enol -P603B |
9 |
28 |
1 |
0 |
6 |
15 |
59 |
10,44 ± 4,16 |
33.36 |
P<0.0001 |
|
Paal2 - r |
21 |
- |
0 |
2 |
- |
7 |
30 |
6.74 ± 4.76 |
21.30 |
P<0.0001 |
|
Paal2 - tl |
21 |
- |
0 |
2 |
- |
7 |
30 |
6.74 ± 4.76 |
21.30 |
P<0.0001 |
|
Paal2 - P603B |
18 |
- |
3 |
2 |
- |
7 |
30 |
16.33 ± 7.50 |
11.43 |
P<0.0010 |