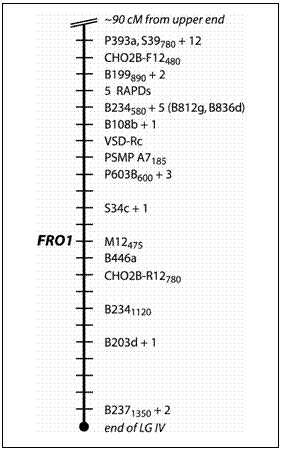
Map position of the FRO1 locus in Pisum sativum
Grusak, M.A., Li, C-M., USDA-ARS Children's Nutrition Res. Center, Dept. of Pediatrics
Baylor College of Medicine, Houston, TX, USA
Moffet, M. and Weeden, N.F Dept. of Plant Sci. and Plant Path.,
Montana State Univ., Bozeman, MT, USA
Iron acquisition in pea, as well as in other dicots and the non-grass monocots, involves a two-step reduction and transport process referred to as Strategy I (8). The root iron reductase in pea [designated FRO1 (9)] is a plasmalemma-localized protein that reduces rhizospheric ferric iron (usually as Fe[III]-chelates) to ferrous iron (Fe2+), prior to the membrane uptake of Fe2+ through the divalent metal transporter, RIT1 (2, 6). Thus, it is an important regulator of whole-plant iron status, especially under alkaline soil conditions where available iron is limiting (8). It also is a potential breeding target for the development of improved cultivars that would be less susceptible to conditions of iron deficiency stress.
Because of our interest in this gene, we wished to map the FRO1 locus and therefore attempted to develop a FRO1-specific CAPS marker that could be used with the JI1794 x Slow mapping population. Over 1,000 segregating markers have been mapped in this recombinant inbred population (1, 11), permitting new markers/loci to be located with high precision. In order to obtain sequences for the FRO1 alleles in both JI1794 and Slow, genomic DNA was extracted from root tissues [hydroponically grown plants (6)] of genotypes Slow and JI1794 using the DNeasy Plant kit (Qiagen, Inc., Valencia, CA, USA). A series of PCR primer sets were then designed from the available PsFRO1 cDNA sequence (Genbank Accession AF405422), and PCR reactions were performed with DNA from both genotypes. The sequencing of a number of overlapping PCR products allowed us to determine the genomic sequences for FRO1 in each parent; these are available as GenBank Accessions AY747697 and AY747696 for genotypes Slow and JI1794, respectively. Although several nucleotide polymorphisms were identified, we selected the region from nucleotides 702 to 1623 in Slow (equivalent to nucleotides 706 to 1623 in JI1794) as a good region for an easily distinguishable CAPS marker. The PCR product in JI1794 contains a polymorphism at nucleotide 1288 (intron) that can be cleaved with EcoRI. Treatment of the PCR products with this restriction enzyme yields a full-length PCR product of 922 nucleotides in Slow and two products of 583 and 335 nucleotides in JI1794.
To map the FRO1 locus in the JI1794 x Slow RIL population, PCR reactions were performed with genomic DNA isolated from 51 F12 individuals. The primers used were: forward: AGAACATGGCCCTAAAGTGTACGTAA; reverse: CCCTTAGAGAAGTTGAGTTCAACGG. Products were treated with EcoRI and separated on 2% agarose gels to score genotypes in each line. The map position of FRO1 also was determined in the F8 RIL population (49 lines) generated from the cross MN313 x JI1794 and in two BC1F6 populations: [Sparkle x WL808] x Sparkle (42 lines) and [Delta x WL808] x Delta (37 lines). In the MN313 x JI1794 population the enzyme Hinf produced a polymorphism, and in the [Delta x WL808] x Delta population, the enzyme Hsp92 II was used to reveal a polymorphism in the gene sequence.
Results
The desired fragment of about 920 bp was amplified in all parents tested (JI1794, WL808, Slow, MN313, Sparkle and Delta). In some amplifications a secondary fragment at about 900 bp was observed after electrophoresis; however multiple fragments were not observed after restriction digests, suggesting that this secondary fragment was an artifact of the electrophoresis process. The EcoR1 site was present in the alleles possessed by JI1794, WL808, MN313 and Delta, whereas alleles in Slow and Sparkle lacked an EcoR1 site. MN313 was found to lack a Hinf site possessed by JI1794, and WL808 displayed a different pattern of Hsp92 II fragments than the other parents.
Segregation at FRO1 did not depart significantly from the expected
1:1 (in the F2-derived RILs) or 3:1 (in the BC-derived RILs) ratios.
In the JI1794 x Slow
RIL population FRO1 mapped about 13 cM distal to
Table
1. Joint segregation analysis of FRO1
with P393 in four pea populations
|
|
|
FRO1,P393 genotype of linea |
Linkage c2 (expected ratio) |
Percent Recomb. |
|||
|
Populationb |
N |
P1P1 |
P1P2 |
P2P1 |
P2P2 |
||
|
JI1794 x Slow |
51 |
23 |
7 |
5 |
16 |
20.3*** (1:1:1:1) |
12 |
|
MN313 x JI1794 |
49 |
18 |
4 |
7 |
20 |
14.9*** (1:1:1:1) |
11 |
|
[D x 808] x D |
37 |
28 |
2 |
4 |
3 |
7.6** (9:3:3:1) |
15 |
|
[Sp x 808] x Sp |
42 |
34 |
3 |
2 |
3 |
13.1*** (9:3:3:1) |
12 |
a P1 = Homozygous for allele from first parent listed, P2 = homozygous for allele from second parent listed ** = P<0.01; *** = P<0.001
b D = Delta, 808 = WL808, Sp = Sparkle

 P393a and
about 6 cM distal to the microsatellite PSMP-A7185 (Table 1; Fig. 1).
This region represents the most distal portion of the lower arm of LGIV
as depicted on the consensus map (11). The
segregation pattern of FRO1 did not display linkage to any other region
of the linkage map. In the remaining
three populations FRO1 also displayed linkage with markers on LG IV
(Table 1). In the MN313
x JI1794 population, FRO1 showed weak linkage with a region on LG V.
This linkage did not increase in intensity in either direction along LG
V, nor was it confirmed in any other population examined.
P393a and
about 6 cM distal to the microsatellite PSMP-A7185 (Table 1; Fig. 1).
This region represents the most distal portion of the lower arm of LGIV
as depicted on the consensus map (11). The
segregation pattern of FRO1 did not display linkage to any other region
of the linkage map. In the remaining
three populations FRO1 also displayed linkage with markers on LG IV
(Table 1). In the MN313
x JI1794 population, FRO1 showed weak linkage with a region on LG V.
This linkage did not increase in intensity in either direction along LG
V, nor was it confirmed in any other population examined.
Discussion
The location of FRO1 near the distal end of the non-satellite arm of LGIV (chromosome 7) makes this gene an important anchor marker on the pea linkage map. Other anchor markers for this region include the isozyme ALATp (11) and three morphological markers, age, wsp and sil (10, 12). Although wsp and sil are excellent markers for crosses in which they segregate, lines possessing these mutations are rarely used as parents to generate populations for mapping or QTL studies. The root mutation, age, is more difficult to score and is not recommended as an anchor marker. Only two alleles at Alatp are known, and one of these is rare (13). In contrast, we identified four distinct alleles of FRO1 in a relatively limited, albeit wide survey of the pea germplasm. Two of these alleles are present in cultivated germplasm (MN313 and Sparkle), suggesting that many other alleles could be identified in a more intensive survey of the gene sequence in other cultivars.
The JI1794 x Slow RIL population possesses the most saturated map of the four populations we used for mapping. FRO1 mapped unambiguously in a cluster of RAPD markers on LGIV, co-segregating with one of them. In the other crosses, FRO1 represented the most distal marker on linkage group IV and did not display tight linkage (<5 cM) with any marker. In all cases the most closely linked anchor marker was P393, approximately 13 cM from FRO1. Although in one population (MN313 x JI1794) FRO1 also exhibited moderate linkage (15-20 cM) to a region on LG V, this linkage was judged to be coincidental because it remained relatively weak regardless of the marker on LGV we tested and failed to be confirmed in any of the other crosses.
Mapping the FRO1 (iron reductase) locus to LGIV is an important step in our continuing investigations into the molecular regulation of plant iron homeostasis. This information will be instructive as we map other iron-related loci in pea, such as those for the ferrous iron transporter, RIT1, or for two iron-hyperaccumulating mutations, brz and dgl. The brz mutant (7), known for bronzing of the leaves, and the dgl mutant (3) known for degenerative leaves, have both been shown to over-accumulate iron due to an up-regulation of root iron reductase activity (5, 6). However, physiological evidence suggests that both mutations induce a shoot-derived, phloem-mobile signal that up-regulates FRO1 and other genes in the roots, rather than being alterations in the FRO1 gene, per se (4, 5). The Brz locus is known to be located at the opposite end of LGIV from FRO1 (11), and it will be interesting to see whether RIT1 and Dgl reside near the FRO1 locus or assort independently.
1. Brauner, S., Murphy, R.L., Walling, J.G., Przyborowski, J. and Weeden, N.F. 2002. J. Amer. Soc. Hort. Sci. 127: 612-622.
2. Cohen, C.K., Fox, T.C., Garvin, D.F. and Kochian, L.V. 1998. Plant Physiol. 116: 1063-1072.
3. Gottschalk, W.G. 1987. Pisum Genetics 19: 9-11.
4. Grusak, M.A. 1995. Planta 197: 111-117.
5. Grusak, M.A. and Pezeshgi, S. 1996. Plant Physiol. 110: 329-334.
6. Grusak, M.A., Welch, R.M. and Kochian, L.V. 1990. Plant Physiol. 94: 1353-1357.
7. Kneen, B.E., LaRue, T.A., Welch, R.M. and Weeden, N.F. 1990. Plant Physiol. 93: 717-722.
8. Marschner, H. and Römheld, V. 1994. Plant Soil 165: 261-274.
9. Waters, B.M., Blevins, D.G. and Eide, D.J. 2002. Plant Physiol. 129: 85-94.
10. Weeden, N.F. 1999. Pisum Genetics 31: 35.
11. Weeden, N.F. 2004. Pisum Genetics 36: 24.
12. Weeden, N.F., Ellis, T.H.N., Timmerman-Vaughan, G.M., Święcicki, W.K., Rozov, S.M. and Berdnikov, V.A. 1998. Pisum Genetics 30: 1-4.
13. Weeden, N.F., Morrell, K and Boone, W.E. 1998. Pisum Genetics 30: 25-26.
14. Weeden, N.F. and Wolko, B. 1988. Measurement of genetic diversity in pea accessions collected near the center of origin of domesticated pea. IPBGR final report, Rome, 20 pp.