PISUM Genetics
2003—Volume 35
Research Papers
Branching in pea: double mutants of rms7
with rms1 through rms5
Murfet, I.C.
Plant Sci, Univ. of Tasmania,
Hobart Tasmania, Australia
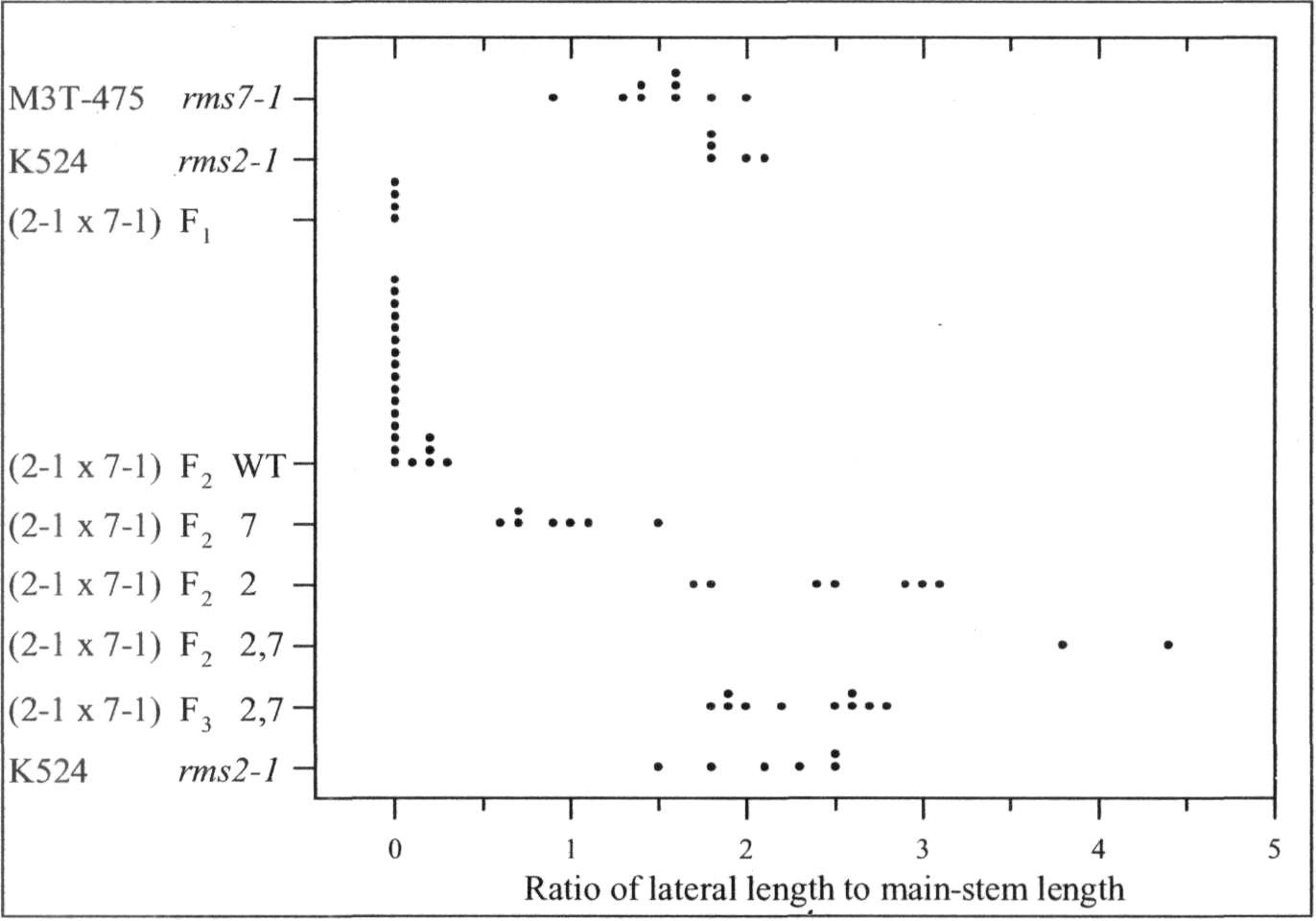
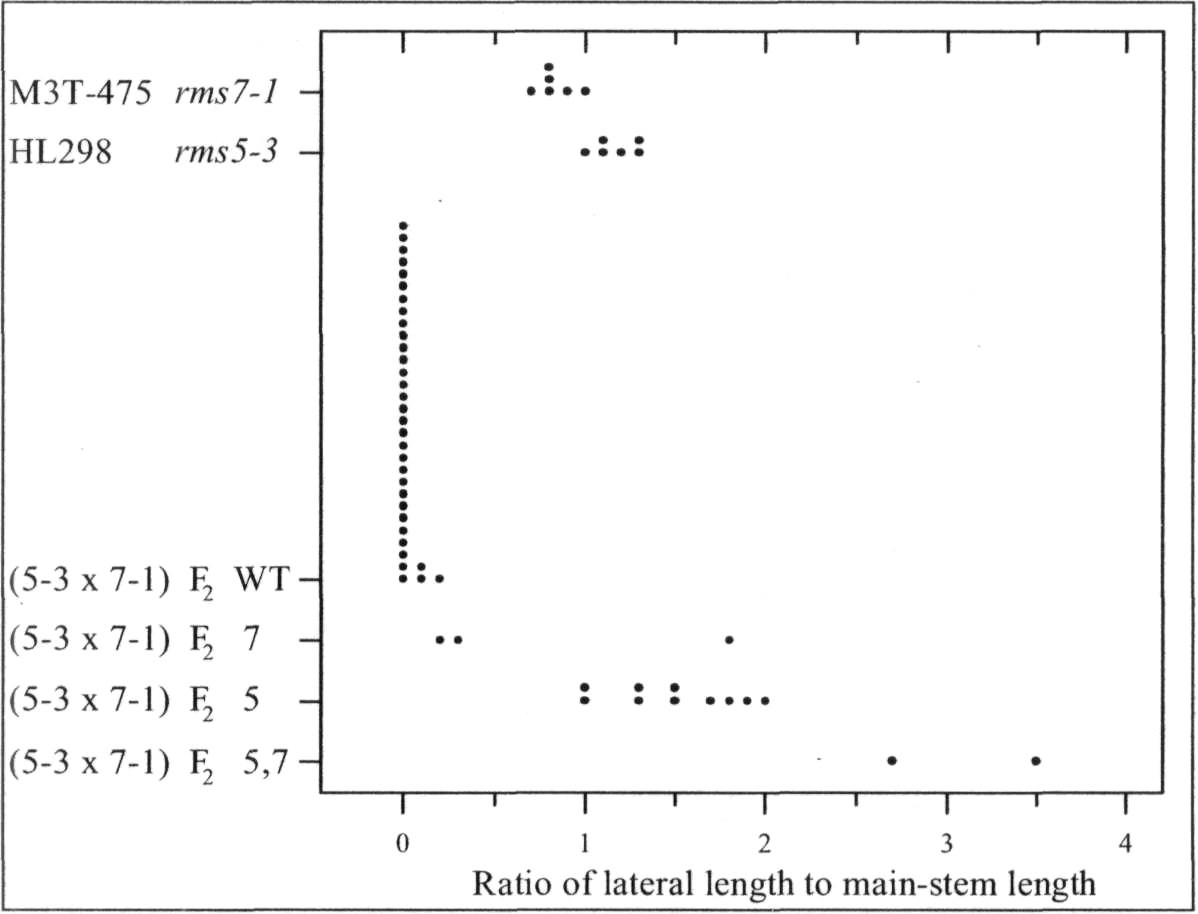
Mutants rms6 and rms7 in pea have increased branching from basal nodes (6, 10). In contrast, mutants rmslthrough rms5 have increased branching from both basal and aerial (upper stem) nodes (1-3). A start has beenmade on checking double-mutant phenotypes. In some cases, the double mutant expresses an additivephenotype with branching more strongly enhanced than in either single mutant, e.g. rms2-1 rms4-1, rms2-1rms5-2, rms3-1 rms6-2 and rms6-1 rms7-1 (6, 8-10). In other cases, epistasis occurs and the double-mutantphenotype does not transgress beyond the range of the single mutants, e.g. rms1-1 rms4-1 and rms2-1 rms3-1 (8).
In the present study, the phenotype of the double mutants of rms7 with rmsl through rms5 was examinedin tall (Le) plants grown in the glasshouse under an 18-h photoperiod (for details see 8). This strategy wasdesigned to allow identification of double-mutant plants regardless of whether they were clearly obvious froman additive double-mutant phenotype or hidden by epistasis of one or other mutant allele. The strategy makesuse of the following information gleaned from years of observation of branching in pea. 1) Basal branching isexpressed more strongly in dwarf (le) than tall (Le) plants (4, 7, 8). 2) In contrast to dwarf plants, tall plantswith WT (wild-type) branching genes invariably fail to produce secondary stems from a basal node under the18-h conditions used. 3) Tall rmsl through rms5 plants always produce aerial laterals under these 18-hconditions. Thus in an F2 population, any tall plant with a major secondary stem and no aerial laterals couldbe considered as homozygous for rms7. In cases where double-mutant plants were not exposed in F2 by anadditive phenotype, F3 progenies could be grown from the homozygous rms7 plants and any F3 plantsexpressing strong growth of aerial
branches would be exposed asdouble mutants.
In accordance with thisstrategy, dwarf line M3T-475(rms7 1) was crossed with tall linesWtl5240 (rms1-5, ex Kaliski),K524 (rms2A, ex Torsdag), K487(rms3-1 ex Torsdag), K164 (rms4-1, ex Torsdag) and HL298 (rms5-3). HL298 was specifically bred forthis purpose from a cross betweentall cv. Torsdag and Wtl5241(rms5-3, ex dwarf cv. Paloma).Further details on these mutantlines are given by Arumingtyas etal. (2).
The rmsl -5 rms7-1 doublemutant was found to have anadditive phenotype (Fig. 1). Tall F2
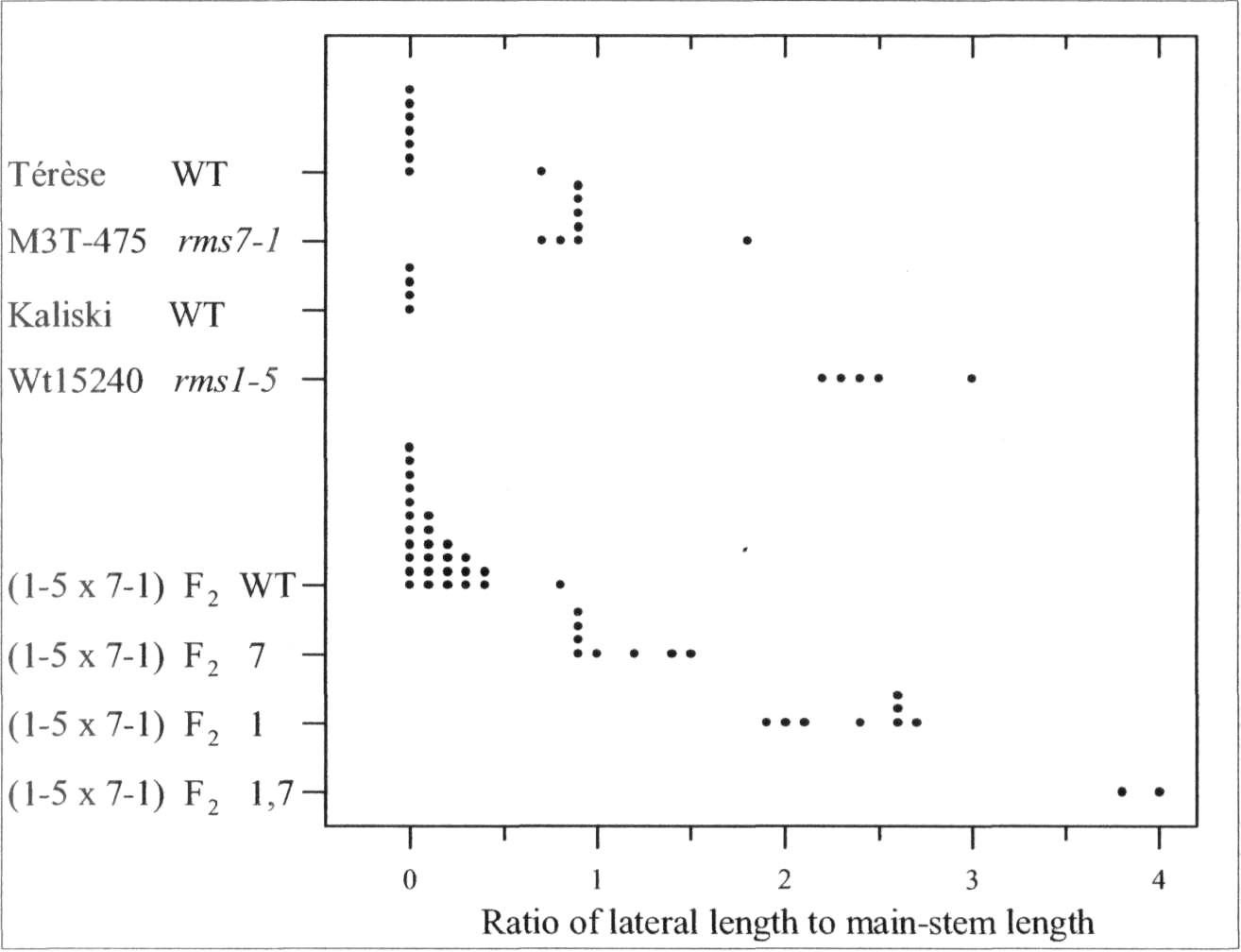
plants of cross Wtl5240 (rms1-5) xM3T-475 (rms7-1) could bepartitioned into four branchingclasses corresponding to WT, rms7,rms1, and rms1 rms7 double-
Fig. 1. Distribution of the branching index 'ratio of lateral to main-stemlength' for cv. Terese, M3T-475 (rms7-1), cv. Kaliski, Wtl5240 (rms1-5), and tall F2 plants from the cross Wtl5240 x M3T-475. The F2 data aresubdivided into four branching phenotypes representing WT, rms7, rms1,and double-mutant rmsl rms7 plants. Data are from mature plants;photoperiod 18 h.