PISUM Genetics
2003—Volume 35
Research Papers
The basal-branching pea mutant rms7-1
Morris, S.E. , Beveridge, C.A.,
Australian Res. Council Cent, of Excellence
Murfet, I.C,
Prioul, S. and Rameau, C.
for Integrative Legume Res., and School of Life Sci. Univ. of Queensland, St Lucia, Australia Plant Sci., Univ. of Tasmania, Hobart, Tasmania, Australia Genetique et Amelioration des Plantes INRA, Versailles, France
Present addr: Tas. Inst, of Ag. Res., PO Box 3523, Burnie, TAS 7320, Australia Corresponding author email: Suzanne.Morris@utas.edu.au
Introduction
The rmsl (ramosus1) through rms5 mutations increase both basal and aerial branching in pea (1,2, 5). In contrast, the rms6 mutation increases basal branching only (12). Between two and eleven mutant alleles have been reported for each of these six RMS loci (12, 16). The ram mutant (9) and the auxin-deficient bsh mutant (17) were also named for their increased branching but their full phenotypic profile distances them somewhat from the rms series of mutants.
Branching habit in pea is also influenced by the background for length and flowering genes: basal branching is enhanced in a dwarf (le) background and under short to intermediate photoperiods in plants with a long-day flowering response (background Sn Dne Ppd) (6). Branching in lupin shows similar trends in response to environmental conditions (8).
Pea mutants rmsl through rms5 have enabled significant progress in our understanding of the control of branching in plants. These rms mutants are the only increased branching mutants to be well characterized for involvement of long-distance signals, known and unknown, in branching control. This characterization was achieved by investigating grafting responses with WT (wild-type) plants, shoot auxin level and transport, auxin responses and root xylem sap cytokinin concentrations. These analyses indicated that two novel graft-transmissible signals are involved in branching control: a feedback signal controlled by RMS2 and a branching inhibitor controlled by RMS1 and RMS5 (4, 7, 10). The hypothesis for branching control has therefore been expanded to incorporate these novel long-distance signals together with the classical phytohormones auxin and cytokinin (3, 10). The recent cloning of RMSJ using the Arabidopsis MAX4 homolog from Medicago truncatula (15) will enable further testing of this hypothesis.
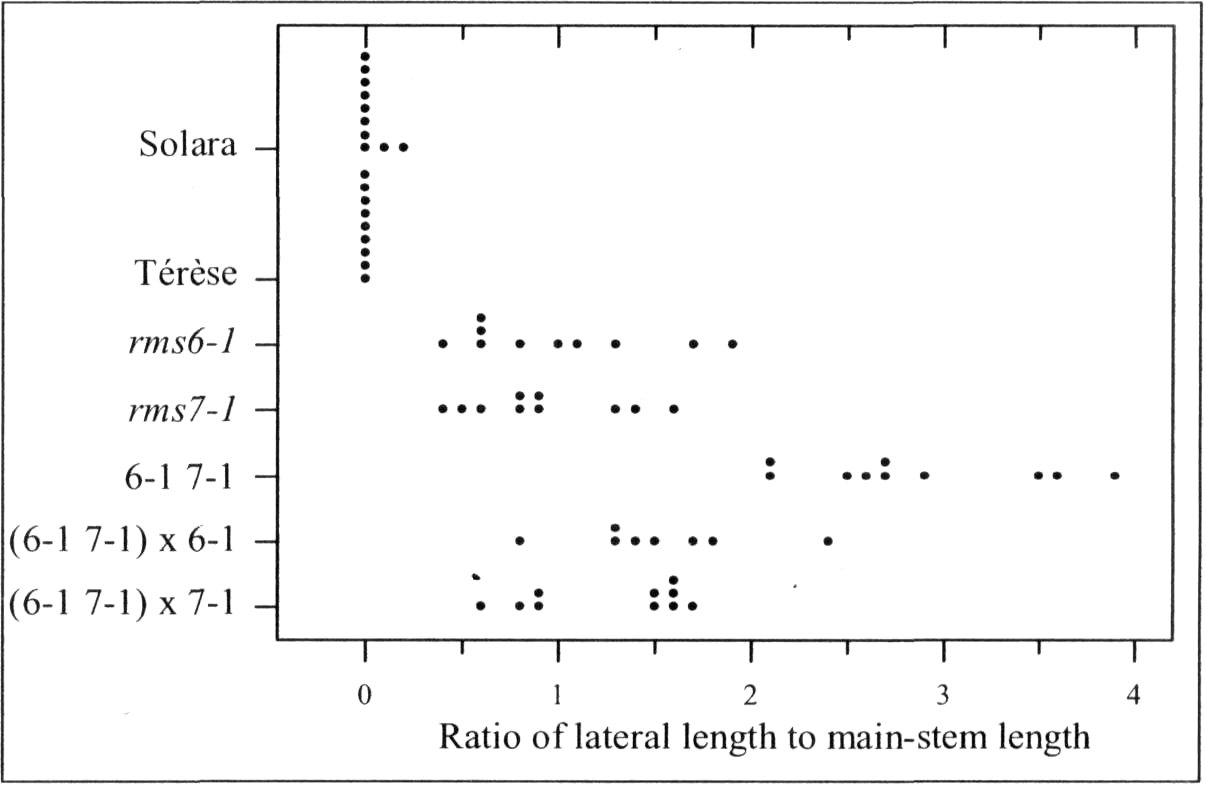
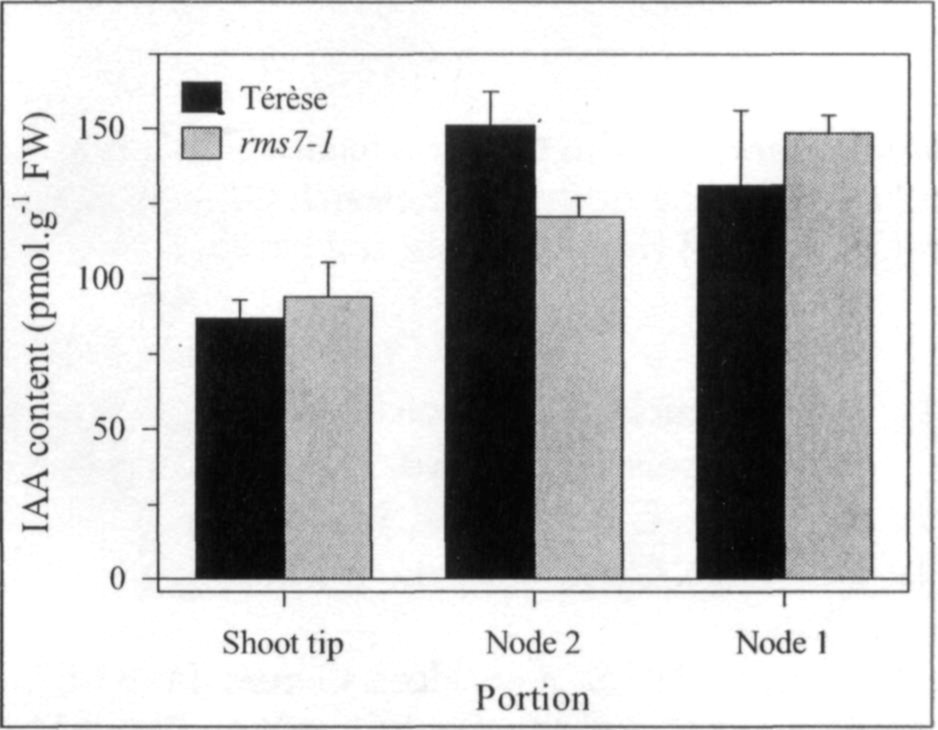
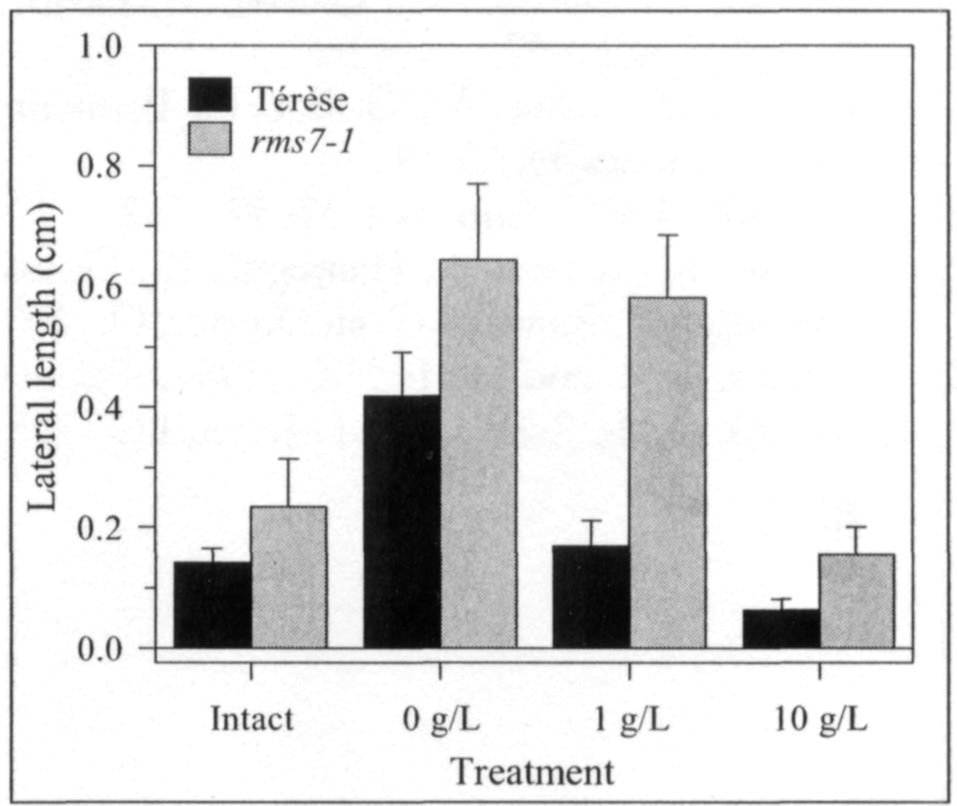
Mutations that specifically enhance basal branching under all photoperiods may be of agronomic value if they increase the number of secondary stems that match the main shoot in vigour and flowering time. Mutants rmsl through rms5 do not fulfil these criteria as both basal and aerial branching is enhanced; their increased aerial branching in comparison with WT is often particularly evident under long-day conditions (2). In contrast, the rms6 mutant plants show increased basal branching under both short and long photoperiods and any tendency to aerial branching is, if anything, diminished in the rms6 mutant compared with WT (11, 12). In this paper we report on a recessive mutant at a new ramosus locus, RMS7, which, like the rms6 mutant, has enhanced branching at the basal nodes only.
The rms7-1 mutant
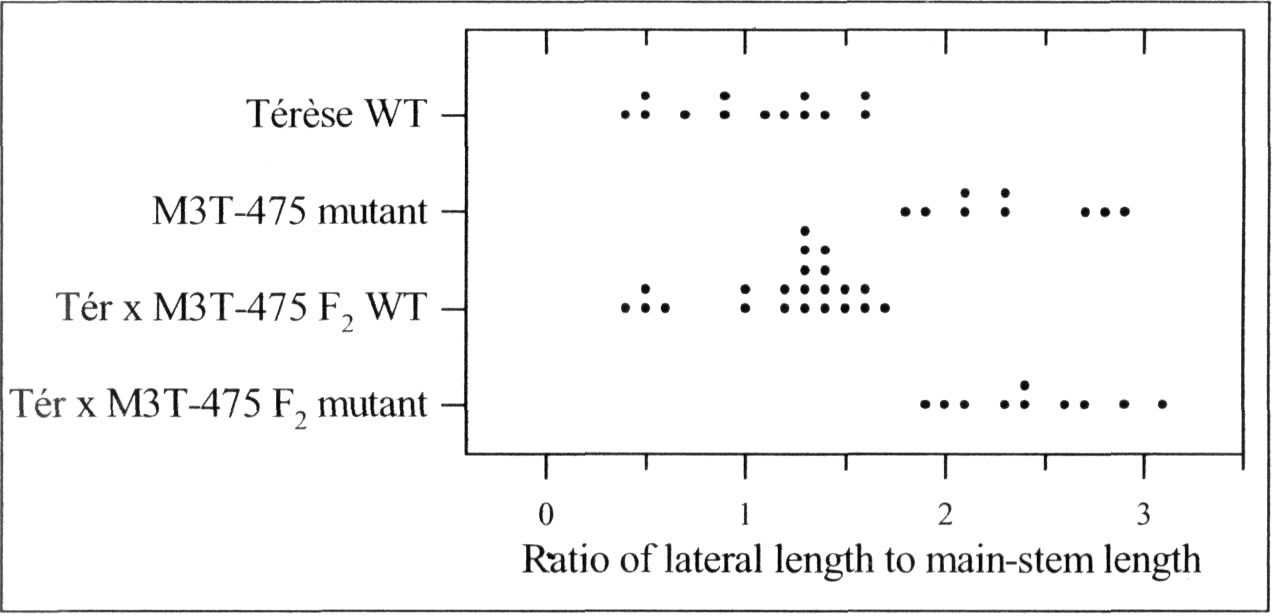
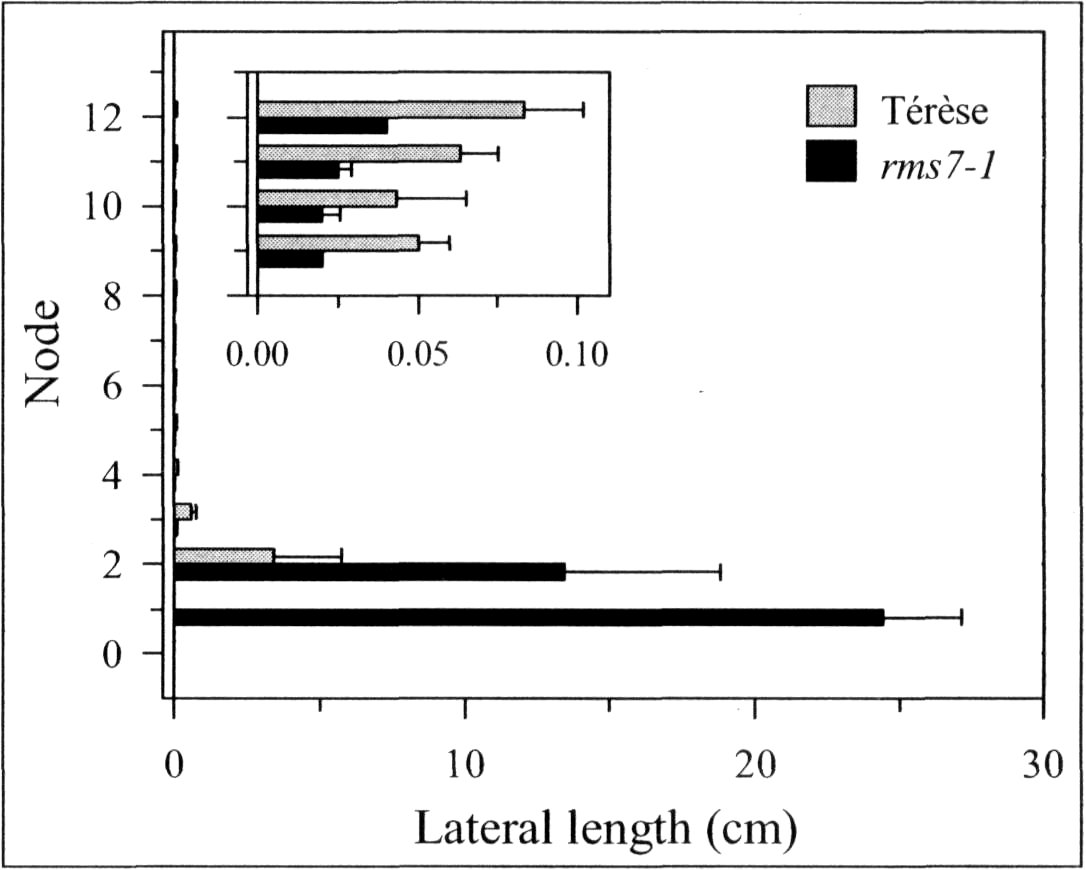
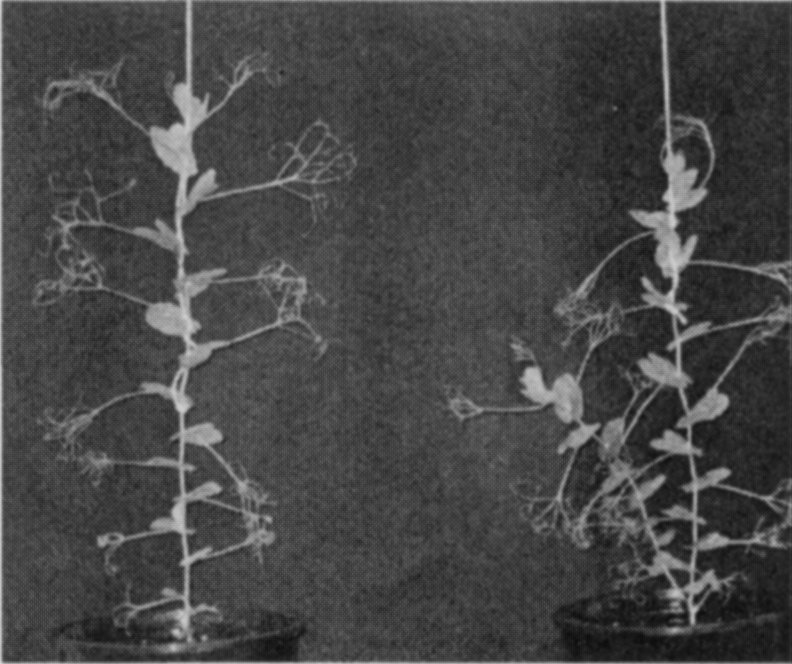
A mutant (M3T-475) with enhanced basal branching (Fig. 1) and recessive inheritance (Fig. 2) was selected at Versailles following EMS mutagenesis of the dwarf, semi-leafless (af), combining cultivar Terese. The M3T-475 mutant
Fig. 1. Branching phenotype of cv. Terese (left) andM3T-475 (rms7-1; right) plants.