Mapping
of two thiamin deficiency genes of pea
Hagh
Nazari, A., Sharma, B. and
Div. of Genetics, Indian Agric. Res. Inst.
Mishra,
S.K.
New Delhi, India
Kumar and Sharma (1) isolated 20 stable auxotrophic mutations for thiamin
biosynthesis in pea. These
mutations were induced by 0.05% ethylmethane sulphonate (EMS) and 0.0057%
nitrosomethyl urea (NMU). Based on
complementation tests, the mutations were recessive alleles (thiA,
thiB, and thiC) at three loci. The
mutants are characterized by yellowing of the leaves about two weeks after
germination, followed by death of the plants.
The mutants were viable and indistinguishable from normal plants when
thiamin was applied exogenously at the concentration of about 0.2 mg/ml.
ThiA
has been mapped very close to K
on linkage group II (10). ThiB
was mapped near Pl in linkage group VI
(11.3 crossover units), and ThiC was
located 20 crossover units away from St
in linkage group III (1). However,
the precise positions of each locus in relation to other loci on the respective
linkage groups have not been studied. The
thiamin auxotrophic mutants can serve as good seedling markers in genetic
studies.
Two of these three mutants, i.e.
thiB (SK 25) and thiC (L 102), are
maintained at the Division of Genetics, I.A.R.I., New Delhi.
In order to determine the precise position of these two genes, the
mutants were crossed with lines having several known markers of linkage groups
III and VI. The data for thiB were obtained
from the cross of SK 25 (thiB) with P
1746-2 (Pl, Arg) while the data for ThiC
were obtained from crosses of line L 102 (thiC)
with P 1297-1 (apu, uni), P 1759-2 (wel),
and P 1563 (st). A cross between the lines P 841 (st) and P 1297-1 was also made to determine the distance between the
loci St and Apu. These crosses also segregated for the genes A,
D, Af, Le, Gp, Tl, Con, and Wlo,
which belong to other linkage groups. The distance between Apu
and Wel was estimated from a cross
between P 1297-1 and P 1759-2. In all the crosses, the F1 plants were
fully fertile. The F1 plants in the crosses involving the thiB
and thiC mutants and normal green parents were normal, confirming the
recessive nature of these two mutations. Details of the crosses, single locus
and joint segregation with c2 values and
recombination fractions (R.F) obtained by using a computer program, CROS,
provided by Dr. S.M. Rozov (http://pisum.bionet.nsc.ru) are presented in Table
1. No mapping function was applied, and the map distances are expressed as
recombination frequencies.
Table
1. Joint F2 segregation
of thiamin-deficient mutations (thiB, thiC)
with other morphological markers.
|
 Loci Loci
|
Phase1
|
Cross
|
Phenotypes2
|
Chi-square
|
Recomb. Fract. + Stan.
Error
|
|
DD
|
Dr
|
rD
|
rr
|
Total
|
Locus 1 (3:1)
|
Locus 2 (3:1)
|
Joint seg.
|
|
thiB
Arg
|
C
|
SKxP 1746-2
|
197
|
26
|
20
|
43
|
286
|
2.343
|
0.116
|
85.95c
|
18.3 + 2.6
|
|
Pl
ThiB
|
C
|
SKxP 1746-2
|
199
|
18
|
24
|
45
|
286
|
0.116
|
1.347
|
88.76c
|
16.5 + 2.4
|
|
Arg
Pl
|
C
|
SKxP 1746-2
|
211
|
06
|
06
|
63
|
233
|
0.116
|
0.115
|
224.20c
|
4.34 + 1.2
|
|
ThiC
Wel
|
R
|
L 102xP 1759-2
|
132
|
59
|
53
|
07
|
251
|
0.160
|
0.224
|
8.80a
|
33.3 + 5.5
|
|
ThiC
Apu
|
R
|
L 102xP1297-1
|
140
|
71
|
66
|
02
|
279
|
0.050
|
0.202
|
25.49c
|
16.6 + 5.7
|
|
ThiC
Uni
|
R
|
L 102xP1297-1
|
148
|
63
|
66
|
02
|
279
|
0.050
|
0.430
|
19.45c
|
18.16 + 5.7
|
|
Uni
Apu
|
R
|
L 102xP1297-1
|
180
|
34
|
26
|
39
|
279
|
0.430
|
0.200
|
49.06c
|
24.67 + 3.0
|
|
Thi
St
|
R
|
L 102xP 1563
|
106
|
52
|
44
|
04
|
204
|
0.320
|
0.529
|
8.40a
|
27.5 + 6.4
|
|
Apu
St
|
R
|
P 1297-1xP 841
|
175
|
78
|
66
|
01
|
320
|
0.090
|
0.016
|
21.30c
|
12.94 + 5.5
|
|
Apu
Wel
|
R
|
P 1297-1xP 1759-2
|
092
|
46
|
58
|
02
|
198
|
0.080
|
0.060
|
22.89c
|
17.90 + 6.8
|
a,
b,
c Mean
P < 0.01, P < 0.001, P = 0.0001, respectively.
1
D = dominant allele and r =
recessive allele
2 C = coupling
and R = repulsion phase linkage
The F2 segregation analysis showed that ThiB
and ThiC as well as all other marker
loci segregated in the 3:1 ratio (Table 1).
In the cross SK 25 x
P 1746-2, thiB was estimated to be
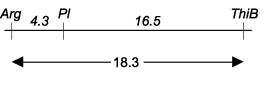
located at about 18.3 map units away from Arg,
and 16.5 map units from Pl. The
linkages ThiB–Arg (P < 0.0001)
and Pl–ThiB (P < 0.0001) were
highly significant. The Pl–ThiB map
distance of 16.5 is reasonably close to 11.3 reported by Kumar and Sharma (1).
The map distance between Pl and Arg
on the same chromosome was 4.3 (Table 1). This
distance was reported as 3 map units by Marx (2), 2 map units by Weller and
Murget (10) and 2.3 map units by Święcicki and Kruszka (9).
Based on the segregation analysis of ThiB, Arg and Pl, these
loci can be arranged in the following order:
From the cross L 102 x
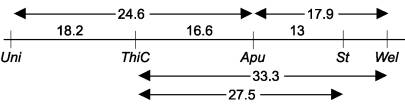
P 1759-2, ThiC has been located at the
distance of 16.6 map units from Apu,
27.5 map units from St, and 33.3 map
units from Wel. Marx (3) located St
about 8.8 map units away from Wel.
Subsequently this distance was reported as 12.5 map units (5).
Sharma and Kumar (8) did not find any linkage between St
and Uni, but this linkage was
estimated to be 36.4 map unitsd by Marx (5).
However, Rozov et al. (6) reported 57 Kosambi units as the distance
between St and Uni.
The results of the cross L 102 x
P 1297-1 showed that ThiC is located
about 18.2 map units away from Uni.
The distance between Uni and Apu being 24.6
shows that ThiC is placed between
these two genes. The linkages ThiC-Apu
and ThiC-Uni were highly significant (P < 0.0001).
Marx (4) reported 23 map units distance between Apu
and Uni, 10 map units between St
and Apu, and 30 map units between
St and Uni. Marx (5) located Apu
at the distance of 33.9 map units from Uni,
while Sarala and Sharma (7) reported 20 map units distance between these two
loci.
From the cross P1297-1 x P841, St
has been mapped 13 map units away from Apu. This distance is reasonably close to 15 map units reported by
Sarala and Sharma (7). Finally,
from the cross P1297-1 x P1259-2, the distance between Wel and Apu was estimated
about 17.9 map units which is very close to the distance (19.8) reported by Marx
(5).
On the basis of joint segregation of thiC,
wel, apu, uni and st, the
following arrangement is proposed:
Thus, our results confirm that the two thiamin auxotrophic mutations, thiB
and thiC, are recessive.
The locus ThiB is located
in the lower part of linkage group VI and ThiC
belongs to linkage group III between Apu
and Uni . The exact position of ThiC
with respect to the isozyme markers (Adh-1
and Acp-3) is under investigation.
The two thiaminless mutations can be used for further studies on the
biochemical pathway of thiamin biosynthesis as well as in mapping programs.
01. Kumar, S. and
Sharma, S.B. 1986.
Mol. Gen. Genet. 204: 473-476.
02. Marx, G.A.
1982. Pisum Newslett. 14: 50-52.
03. Marx, G.A.
1983. Pisum Newslett. 15: 47.
04. Marx, G.A.
1984. Pisum Newslett. 16: 46-48.
05. Marx, G.A.
1986. Pisum Newslett. 18: 49-52.
06. Rozov, S.M.,
Borisov, A.Y. and Tsyganov, V.E. 1994.
Pisum Genetics 26: 24-25.
07 Sarala,
K. and Sharma, B. 1994.
Pisum Genetcs 26: 28.
08. Sharma, B. and
Kumar, S. 1981.
Pulse Crops Newslett. 1(3): 21-22/
09. Święcicki,
W.K. and Kruszka, K. 1995.
Pisum Genetics 27: 22.
10.
Weller, J.L. and Murfet, I.C. 1994.
Pisum Genetics 26: 41-43.
![]() Loci
Loci

