A gene influencing tolerance to common root rot
is located on linkage group IV
Weeden,
N.F.
Dept. of Plant Sciences and Plant Pathology
Montana State University, Bozeman, MT
McGee,
R.
Green Giant Research Center, LeSueur, MN
Grau,
C. R. and
Dept. of Plant Pathology, University of Wisconsin, Madison, WI
Muehlbauer,
F. J.
USDA ARS, Washington State University, Pullman, WA
Breeding for tolerance to common root rot, caused by the pathogen Aphanomyces
euteiches, has been difficult because of the polygenic nature of the
tolerance and the strong influence of the environment.
Crosses between tolerant and susceptible lines or between different
tolerant lines often produce progeny with little or no tolerance (J. Kraft,
personal communication). In the last few years we have concentrated on investigating
the genetic basis of tolerance in MN313, a line developed by Dr. D. Davis at the
University of Minnesota. Further
selection of this release was performed by one of the current authors (C.R.G.).
This line displays good tolerance (scores of 1.5 to 2.0 on a 5 point
scale, where 1.0 indicates no evidence of the disease and 5.0 indicates plant
death before pod set) in fields infested with common root rot in Minnesota and
Wisconsin.
Materials and Methods
Our approach has been to make crosses between MN313 and lines possessing
markers or other traits of interest and to produce populations of recombinant
inbred lines (RILs), with each RIL derived from a separate F2 plant
through single seed descent. One
such cross was made with OSU1026, a release from the breeding program of Dr. Jim
Baggett at Oregon State University. OSU1026
was a dwarf, white-flowered, multiple disease resistant release, containing sbm1,
En, er1, mo,
and Fw. To our knowledge
OSU1026 was never selected for tolerance to common root rot.
MN313 lacked resistance to powdery mildew, and the goal of the cross was
to introgress powdery mildew resistance (er1)
into a line with good tolerance to common root rot.
However, the RIL population proved to be very useful for identifying a
gene in MN313 contributing to tolerance to common root rot.
The MN313 x OSU1026 RIL population consisted of 45 RILs.
We grew the population in the common root rot nursery at Le Sueur, MN as
an F5 in 1999 and an F6 in 2000.
We also grew the F6 in the common root rot nursery at the
Spillman Farm, Pullman, WA in 2000. In
each case at least 10 seeds per RIL were planted. The plants were scored twice during the growing season for
general vigor and symptoms of common root rot.
Plants were scored for disease severity on a scale of 1 to 5 as described
above.
A linkage map was constructed for the population using classical markers,
isozymes and DNA polymorphisms (STS markers and RAPDs).
Possible relationships between markers and disease response were tested
by one way ANOVA calculations.
Results
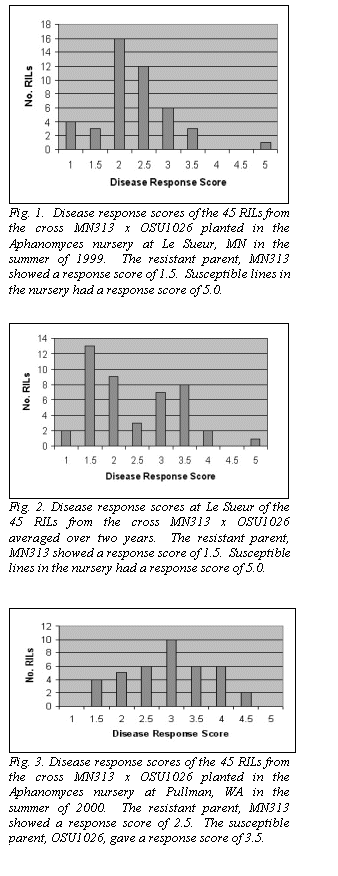
In
1999 at Le Sueur the MN313 parent was given a 1.5 rating and all susceptible
controls (Sparkle, A1078-239, Badger) were scored as 5.0.
OSU1026 was not tested due to insufficient seed available.
All except one of the MN313 x OSU1026 RILs displayed some tolerance
(score of 3.5 or better), with 22 of the 45 lines scoring 2.0 or better (Fig.
1). The one line that was scored as
susceptible (5.0) was located at the end of a row and may have been subjected to
other environmental effects.
In 2000 at LeSueur, a similar pattern was observed in that approximately
half the lines were rated as good or better than the MN313 parent.
The other half of the lines ranged from moderately tolerant to
susceptible (2.5 to 4.0). The
results for individual lines were highly consistent between years, although
those lines rated slightly tolerant in 1999 often displayed more severe symptoms
in 2000. Again the susceptible
controls all were rated between 4.0 and 5.0.
The distribution of two-year averages at Le Sueur for each line is given
in Fig. 2. The distribution is
clearly bimodal, suggesting the influence of a single gene segregating in a 1:1
ratio. Joint segregation analysis
of genes or markers segregating in the RILs and common root rot resistance at Le
Sueur failed to reveal a correlation with er1,
sbm1, or DNA markers on linkage groups
II, VI or VII. However, a significant correlation (p<0.0001) was found
between the root rot resistance phenotype and markers on linkage group IV (P393
and PgmF390). The mean
score for lines with the MN313 allele was 1.85+0.11, while that for lines
with OSU1026 allele was 3.27+ 0.13.
 The results from Pullman were significantly different. Both parents expressed modest tolerance (2.5 to 3.5).
The RILs appeared to segregate for tolerance, but few lines were highly
tolerant or highly susceptible (Fig. 3). There
was little correlation between the results at Pullman and those at Le Sueur. No correlation was observed between linkage group IV markers
and root rot tolerance at Pullman. More
generally, we could not find an association between the segregation of plant
phenotype (disease rating score) at Pullman and that for any of the markers we
have scored in this population. However,
significant gaps still exist in our linkage map for this population, and a lack
of correlation at this point does not necessarily indicate that the variation in
disease response at Pullman lacks a genetic component.
The results from Pullman were significantly different. Both parents expressed modest tolerance (2.5 to 3.5).
The RILs appeared to segregate for tolerance, but few lines were highly
tolerant or highly susceptible (Fig. 3). There
was little correlation between the results at Pullman and those at Le Sueur. No correlation was observed between linkage group IV markers
and root rot tolerance at Pullman. More
generally, we could not find an association between the segregation of plant
phenotype (disease rating score) at Pullman and that for any of the markers we
have scored in this population. However,
significant gaps still exist in our linkage map for this population, and a lack
of correlation at this point does not necessarily indicate that the variation in
disease response at Pullman lacks a genetic component.
Discussion
Our results reveal an important locus affecting the susceptibility of a
pea plant to common root rot in the Aphanomyces nursery at Le Sueur, MN.
The effect was consistent over two years although in the first year the
bimodal pattern was not as obvious. Apparently
the greater severity of the disease in summer 2000 caused the more susceptible
genotype to display more prominent symptoms and thus become a distinct group in
the analysis (Fig. 2).
Two major conclusions can be made from the results presented.
One is that a gene in MN313 located on linkage group IV near P393 has a
significant influence on the expression of tolerance to common root rot in the
field at Le Sueur. This gene
represents the major genetic difference between MN313 and OSU1026 in their
respective responses to the pathogen. In
a different RIL population, derived from the cross MN313 x A1078-239, we have
obtained evidence that this same gene is segregating and influencing Aphanomyces
tolerance in the field at Le Sueur (data not presented).
However, the effect is less clear because A1078-239 is susceptible
(response score 5.0), and we have yet to tag and locate the other gene(s)
affecting tolerance that appears to segregate in this population.
MN313 and OSU1026 must share these other genes bestowing tolerance to
common root rot.
The second conclusion is that the pathogen or set of pathogens in the Le
Sueur field is different from that at Pullman.
Mn313 exhibited high levels of tolerance both years at Le Sueur and was
clearly superior to Sparkle or A1078-239. At
Pullman MN313 displayed little tolerance and was not rated above either of the
susceptible controls. The reverse
situation has been observed in experiments on RILs derived from crosses using
common root rot tolerant lines developed by John Kraft.
In this case Kraft’s lines were definitely superior at Pullman but not
at Le Sueur (Clare Coyne, personal communication).
This conclusion is supported by the work of Malvick and Percich (1) who
demonstrated significant phenotypic differences between strains of A. euteiches sampled from
Minnesota and Oregon. We are
investigating the nature of the pathogens in both fields.
Small quantities of seed are available for several of the MN313 x OSU1026
RILs that combine resistance to common root rot at Le Sueur with powdery mildew
resistance. Breeders and
researchers interested such seed should contact the senior author.
Acknowledgement: This work was supported by grants from the Cool Season Food
Legume Research Committee.
1.
Malvick, D.K. and Percich, J.A. 1998.
Phytopathology 88: 915-921.
 The results from Pullman were significantly different. Both parents expressed modest tolerance (2.5 to 3.5).
The RILs appeared to segregate for tolerance, but few lines were highly
tolerant or highly susceptible (Fig. 3). There
was little correlation between the results at Pullman and those at Le Sueur. No correlation was observed between linkage group IV markers
and root rot tolerance at Pullman. More
generally, we could not find an association between the segregation of plant
phenotype (disease rating score) at Pullman and that for any of the markers we
have scored in this population. However,
significant gaps still exist in our linkage map for this population, and a lack
of correlation at this point does not necessarily indicate that the variation in
disease response at Pullman lacks a genetic component.
The results from Pullman were significantly different. Both parents expressed modest tolerance (2.5 to 3.5).
The RILs appeared to segregate for tolerance, but few lines were highly
tolerant or highly susceptible (Fig. 3). There
was little correlation between the results at Pullman and those at Le Sueur. No correlation was observed between linkage group IV markers
and root rot tolerance at Pullman. More
generally, we could not find an association between the segregation of plant
phenotype (disease rating score) at Pullman and that for any of the markers we
have scored in this population. However,
significant gaps still exist in our linkage map for this population, and a lack
of correlation at this point does not necessarily indicate that the variation in
disease response at Pullman lacks a genetic component.