Densinodosum - a new gene on linkage group III
Swiecicki, W.K. and Wolko, B. Institute of Plant Genetics, Polish Academy of Sciences,
Strzeszynska 34, 60-479 Poznan, Poland
Among numerous mutation types following treatment of dry seeds of Wt 3527 (cv. Paloma) with a combined dose 200 r Nf + 0.014 % NEU the densinodosum mutant gene was selected in the M2 generation (2). After multiplication the line was included to the Pisum Gene Bank at Wiatrowo with the catalogue number Wt 11242. The name densinodosum is justified by a phenotype—mutant plants in comparison to the initial line have shorter stem with a larger number of nodes (Table 1). Number of nodes to the first flower remains unchanged.
Table 1. Selected, morphological characters of densinodosum mutant plants and initial line Wt 3527.
|
Line
|
Stem length (cm)
|
Number of nodes
|
Number of nodes to the 1st flower
|
|
|
|
|
|
|
Wt 3527 (Paloma) -initial |
41.0 |
18.2 |
13.8 |
|
Wt 11 242 (dnd – mutant) |
33.1 |
21.5 |
13.9 |
|
|
|
|
|
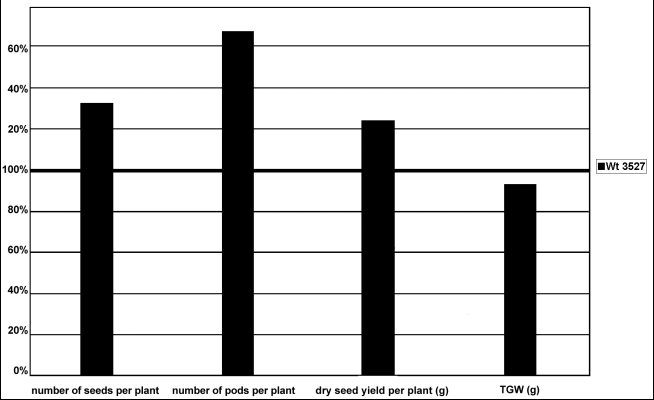
After multiplication of mutant plants, aspects of dry seed yield were also compared (Fig. 1). It appeared that the densinodosum mutant produced higher yield thanks to a larger number of pods and seeds per plant. Thousand grain weight is smaller.

Fig. 1. Aspects of dry seed yield of densinodosum mutant plants related to the initial line Wt 3527 = 100%.
The locus identity test cross for the densinodosum and two genes with a similar phenotype, art1 and art2, was conduced (3). F1 plants of all cross combinations were normal, suggesting that densinodosum (Wt 11242) and arthritic mutants (Wt 16126 - art1 and Wt16125 - art2) are not allelic.
For mapping the new mutant gene the line Wt 11242 was crossed with the set of tester lines with markers for seven linkage groups. An analysis of a monohybrid segregation in F2 generations showed Mendelian, recessive nature of the new gene (Table 2.). The symbol dnd is suggested. Undisturbed, monohybrid segregation of most of observed markers was also observed.
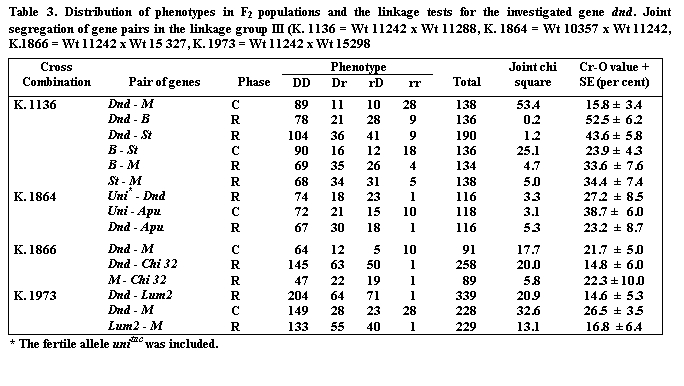
Table 2. Monohybrid segregation for the investigated gene dnd and gene markers in the linkage group III observed in F2 populations of the following linkage test crosses: K. 1140 = Wt 11242 x Wt 11540; K. 1139 = Wt 11242 x Wt 11538; K. 1137 = Wt 11242 x Wt 11777; K. 1135 = Wt 11242 x Wt 11238; K. 1136 = Wt 11242 x Wt 11288; K. 1864 = Wt 10357 x Wt 11242; K. 1866 = Wt 11242 x Wt 15327; K. 1973 = Wt 11242 x Wt 15298.
|
Cross combination |
Gene |
Allele |
Total |
Chi square* (3:1) |
|
|
dominant |
recessive |
||||
|
K. 1140 |
Dnd |
80 |
27 |
107 |
0.00 |
|
K. 1139 |
Dnd |
80 |
25 |
105 |
0.08 |
|
K. 1137 |
Dnd |
84 |
19 |
103 |
2.36 |
|
K. 1135 |
Dnd |
107 |
36 |
143 |
0.00 |
|
K. 1136 |
Dnd |
140 |
50 |
190 |
0.18 |
|
K. 1864 |
Dnd |
97 |
19 |
116 |
4.60 |
|
K. 1866 |
Dnd |
211 |
52 |
263 |
3.83 |
|
K. 1973 |
Dnd |
268 |
71 |
339 |
2.97 |
|
K. 1136 |
B |
106 |
30 |
136 |
0.63 |
|
|
St |
148 |
47 |
195 |
0.08 |
|
|
M |
99 |
39 |
138 |
0.78 |
|
K. 1864 |
Uni |
93 |
25 |
118 |
0.92 |
|
|
Apu |
87 |
31 |
118 |
0.10 |
|
K. 1866 |
M |
69 |
22 |
91 |
0.03 |
|
|
Chi 32 |
197 |
64 |
261 |
0.03 |
|
K.1973 |
Lum2 |
285 |
65 |
350 |
7.71 |
|
|
M |
173 |
56 |
229 |
0.04 |
* c2 = 3.84
Observations of plants in F2 generations gave undisturbed dihybrid segregations for gene pairs dnd—gene marker for linkage groups I, II, IV, V, VI and VII. Disturbed, dihybrid segregation was found in the cross combination K. 1136 for dnd-M from linkage group III with Cr-O value 15.8 (Table 3). However, dnd did not show linkage with either b or st in K. 1136 or with unitac or apu in the cross combination K. 1864.
Two chlorophyll genes–chi32 and lum2–were previously mapped in our laboratory near M on linkage group III (1, 4). Therefore the type line for dnd was crossed to the lines Wt 15327 (chi32, M) and Wt 15298 (lum2, M). Dnd showed clear linkages with both Lum2 and Chi32, confirming its location in the M-segment. At this stage of mapping studies it is impossible to state on which side of M Dnd is located, although the absence of linkage with Uni suggests that Dnd may be on the opposite side of M from Uni.
1. Czerwinska, St. and Wolko B. 1991. Pisum Genetics 23: 14-15.
2. Swiecicki, W.K. 1984. PNL, 16: 84-86.
3. Swiecicki, W.K. 1986. Genet. Pol. 27: 73-80.
4. Swiecicki, W.K. and Wolko, B. 1991. Pisum Genetics 23: 40-41.