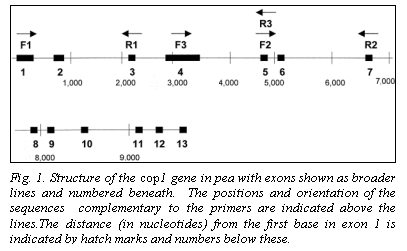
Location and STS primers for Cop1
Murphy, R.L., Weeden, N.F. and Dept. Plant Sciences and Plant Pathology
Montana State University, Bozeman, MT
Przyborowski, J.A. Dept. Plant Breeding and Seed Production
University of Warmia & Mazury, Olsztyn-Kortowo, Poland
The pea mutant lip (light-independent photomorphogenesis) has recently been cloned and shown to be caused by a duplication in the gene homologous to cop1 in Arabidopsis thaliana (3). The mutant was originally characterized by Frances et al. (2) in an Alaska line that failed to display normal etiolation response. Instead, homozygous recessive plants develop expanded leaflets and stipules (although these usually remain yellowish) and stems remain relatively short (approximately 0.6X the height of the wild type) when grown in the absence of light. One of us (NFW) has made several unsuccessful attempts at mapping the mutation, primarily due to difficulties in scoring the mutant phenotype in populations segregating for various marker genes. With the determination of the genomic sequence of the gene (3), we decided to pursue the mapping of the gene using the sequence tagged site (STS) approach described recently (5).
Materials and Methods
Three populations were used in the mapping analysis: JI1794 x Slow recombinant inbred lines (RILs), MN313 x JI1794 RILs and Lip x JI1794 F4s. The JI1794 x Slow RILs represent our standard mapping population that was used to develop the backbone for the consensus map (4). The second set of RILs (MN313 x JI1794) are currently in the F6 generation, and an extensive linkage map has also been developed for this population. The third population is relatively small (derived from 20 F2 plants) but was carefully scored for segregation for lip in the F3.

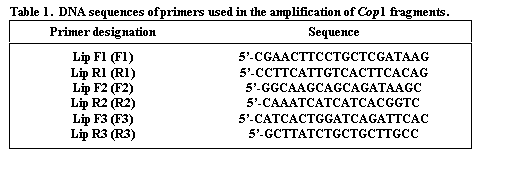
The genomic coding sequence of Cop1 in pea spans nearly 10,000 nucleotides, with 13 exons (Fig. 1), providing ample opportunity to identify a polymorphic region. The lip mutation is produced by a 7700 bp duplication of a section of the promotor region and coding sequence. We selected three regions of the gene for analysis (Fig. 1). The sequences for the primers (Lip F1, Lip F2, Lip F3, Lip R1, Lip R2 and Lip R3) are given in Table 1. Amplification conditions for Lip F1 + Lip R1 (F1R1) and Lip F2 + Lip R2 (F2R2) were 98oC for 30 sec (initial denaturation) followed by 30 cycles of 94oC (10 sec), 60oC (1 min), and 72oC (3 min). A final extension at 72oC for 10 min concluded the amplification process. For the primer set Lip F3 + Lip R3 (F3R3) different cycle conditions were used: 94oC for 2 min followed by 35 cycles of 94oC (1 min), 60oC (2 min), and 72oC (2 min). The final extension at 72oC was for 8 min. For F1R1 and F2R2 amplifications Accutaq LA DNA polymerase (Sigma) was used as per instructions supplied with the product. For F3R3 the amplification mix contained (per tube) 0.1 ul Taq polymerase (Promega), 2.5 ul 10X buffer (Promega), 3.0 ul 0.1 M MgCl2, 0.62 ul of a solution containing 10mM each of each dNTP, 0.25 ul of 20 mM F3, 0.25 ul of 20 mM R3 and 1.0 ul of genomic DNA.
Results
Initial amplification using the F1R1 or F2R2 primer combinations and Taq polymerase from Promega under routine conditions gave inconsistent results due to the length of the amplified product. We therefore used Sigma Accutaq LA DNA polymerase and followed the conditions recommended by the manufacturer. This modification permitted resulted in consistent amplification of the large fragments. Simultaneously, we designed the F3R3 primers to amplify a shorter segment.
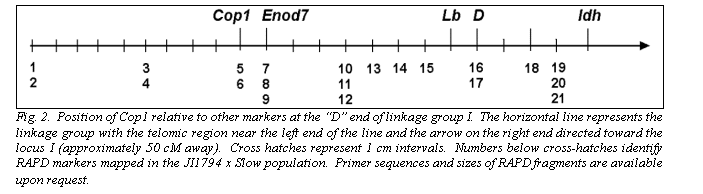
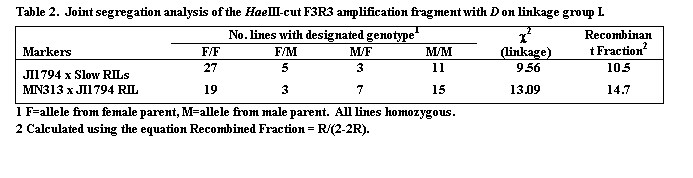
The segregation patterns of HaeIII restricted F1R1, F2R2 and F3R3 fragments in the JI1794 x Slow RILs were all identical, confirming that all three fragments are derived from the Cop1 genomic sequence. Comparison of this segregation pattern with those for the over 1,000 markers mapped in this population placed Cop1 near the end of linkage group (LG) I distal to the locus D (Table 2, Fig. 2). This position was confirmed in the MN313 x JI1794 population (Table 2). On both maps Cop1 was placed about 10 cM from D. The position of Cop1 was unambiguous in that Cop1 failed to display significant linkage with any other region of the genome in either population.
Confirmation that the Cop1 locus cosegregates with the lip mutation was obtained in the third cross. Although markers were not scored in this cross, five F4 homozygous lip lines all contained the Alaska allele at Cop1 and four true breeding wild type lines all possessed the JI1794 allele. As expected, F4 plants derived from F3 families segregating for lip showed one of the parental phenotypes at Cop1 or were heterozygous.
Discussion
The location of Cop1 near the top of LG I provides a very useful STS marker for this region of the pea genome. Other loci approximately 10 cM distal of D include Sym2, Nod3 and a cDNA (c44) described in Ellis et al. (1). At present, Cop1 appears to be the most convenient of these loci for marking the region and mapping other genes. However, the level of polymorphism present within pea for the genomic sequence of Cop1 has yet to be thoroughly tested. Digestion of the F3R3 product from several domesticated lines with several restriction enzymes suggests that this fragment displays relatively little polymorphism within commercial germplasm (R.L. Murphy, unpublished results). Studies on the F1R1 and F2R2 amplification products have yet to be performed. For comparison between genera, preliminary evidence indicates that at least the F2R2 combination amplifies a homologous sequence in lentil, permitting mapping of the Cop1 locus in a related legume (J.G. Walling, R.L. Murphy, and N.F. Weeden unpublished). Thus, the approach to mapping genes in pea that we have taken in this study should eventually provide a set of primers that can be used for comparative mapping in a wide selection of legumes.
1. Ellis, T.H.N., Turner, L., Hellens, R.P., Lee, D., Harker, C.L., Enard, C., Domoney, C. and Davies, D.R. 1992. Genetics 130: 649-663.
2. Francis, S., White, M.J., Edgerton, M.D., Jones, A.M., Elliott, R.C. and Thompson, W.F. 1992. Plant Cell 4: 1519-1530.
3. Sullivan, J.A. and Gray, J.C. 2000. GenBank accession AJ276592.
4. Weeden, N.F., Ellis, T.H.N., Timmerman-Vaughan, G.M., Swiecicki, W.K., Rozov, S.M. and Berdnikov, V.A. 1998. Pisum Genetics 30: 1-4.
5. Weeden N.F., Tonguc, M. and Boone, W.E. 1999. Pisum Genetics 31: 30-32.