Construction of a bacterial artificial chromosome
library
for pea (Pisum
sativum L.)
Coyne, C.J.,
USDA-ARS Plant Introduction
Washington State University, Pullman, WA 99164-6402
Meksem K., Lightfoot, D.A.,
Dept. of Plant, Soil, and General Agriculture
Southern Illinois University, Carbondale, IL 62901‑4415
Keller, K.E.,
Martin, R.R.,
USDA-ARS Horticulture Crops Research Laboratory
Corvallis, OR 97330
McClendon, M.T.,
Dept. of Horticulture, Washington State University
Pullman, WA 99164-6402
Inglis, D.A.,
Mount Vernon Research and Extension
Washington State University, Mount Vernon, WA 98273-9761
Storlie, E.W.
and
Department of Biological & Physical Sciences
Univ. of Southern Queensland, Toowoomba, Queensland 4350, Australia
McPhee, K.E.
USDA-ARS Grain Legumes, Dept. of Crop Science
Washington State University, Pullman, WA 99164-6402
Introduction
One of the major constraints to fully utilizing the U.S. plant germplasm
resources is the paucity of effective techniques to efficiently discover new
alleles useful to crop breeding (12). Several
key genomic technologies relevant to germplasm collections, however, have been
developed (1). Information gathered
using this functional genomics approach may be useful in identifying disease and
stress resistance alleles in U.S. pea germplasm accessions as a molecular tool
for efficient curation of the collection. Because
the USDA-ARS National Plant
Germplasm System (NPGS) encourages its system of repositories to embrace new
technologies to maximize the conservation of genetic variation for economically
important traits in each collected crop species, one such biotechnology tool, a
bacterial artificial chromosome (BAC) library of the pea genome, has the
potential to provide new opportunities for the development and management of pea
genetic resources.
Our objective was to construct a bacterial artificial chromosome (BAC)
library of Pisum sativum from the
germplasm line PI 269818 of the NPGS in the binary vector pCLD04541, (V41).
The binary vector potentially allows for the direct transformation of
candidate pea gene BACs into plants to verify the phenotype (11).
Pea has a number of stable and single gene traits useful to breeders,
including resistance to pea seedborne mosaic virus (PSbMV) conferred by sbm-1.
PSbMV has been disseminated worldwide in infected seed (7).
The NPGS places a high priority to maintain and distribute Pisum
germplasm tested free of seed-borne pathogens such as PSbMV.
Thus, virus-free seed is critical to the mission of the cool season food
legume program of the Western Regional Plant Introduction Station at Washington
State University. The library will
be useful for (i) positional cloning of disease resistance genes, such as sbm-1
and other economic traits (2), (ii) targeted microsatellite development to
economic traits of interest (4, 9), and (iii) use of the microsatellite markers
for fingerprinting PI accessions to eliminate duplication of pea germplasm and
assess genetic diversity in the collection (5).
Materials and methods
Plant
materials
Seed of PI 269818 was obtained from the National Plant Germplasm System.
For DNA isolation, plants were grown in a growth chamber in soiless
mixture for 14 days, with 16 h light, 8 h dark at 22°C. Two weeks after
germination, seedlings were grown in continuous dark for 3 days to reduce the
carbohydrate content of the leaves.
Vectors
and DNA preparation
The BAC vector V41 (pCLD04541) was developed by Dr. J.D.G. Jones (8) and
generously provided by Dr. H. Zhang (Texas A & M University, USA). The V41
is a binary vector for Agrobacterium-mediated
plant transformation and has been shown to be capable of stable maintenance of
large plant DNA (9, 11). The method
for high molecular weight (HMW) DNA preparation from plant nuclei was described
by Zhang et al. (13). Nuclei were prepared from 25 g of leaves and embedded in
12 ml of 0.5% (w/v) low melting point agarose plugs.
The plugs were cut into 12
pieces, partially digested for 10 min at 37°C with HindIII (Gibco-BRL, MD) and BamHI
(Gibco-BRL, MD). Restriction enzyme
concentration was optimized for each set of HMW DNA isolations.
Test digests were performed in seven increment steps of 0.3 to 5 units of
enzyme per 1 mg
of DNA followed by separation using PFGE (pulsed field gel electrophoresis) on a
CHEF DRIII (Bio-Rad, Hercules, CA).
Each digestion was carried out in a 200 ml total volume. Reactions were
stopped by adding 20 ml of ice cold 0.5 M EDTA pH 8, on ice.
Construction
and storage of the BAC library
The vector V41 was purified with Qiagen plasmid purification kit,
followed by two cesium chloride gradients. The vector was digested (HindIII
or BamHI ) and then dephosphorylated
to prevent self ligation. Partially digested HMW DNA was size selected on 1%
(w/v) pulsed field low melting point agarose gels in 0.5x TBE (45 mM Trizma
base, 45 mM boric acid and 1 mM EDTA, pH 8.3) by PFGE. Two size-selections were
performed to increase the average insert size of the BACs, and to eliminate
small DNA fragments trapped in the HMW fraction. The first PFGE size selection
was performed for 18 h at 11°C with constant pulse time of 90 s, at a 120°
angle and 6 V/cm. The second size-selection was performed for 12 h at 11°C with
an initial pulse time of 1 s, a final pulse time of 10 s, at a 120° angle and 6
V/cm.
After the second PFGE, two methods were used to isolate the
size‑selected DNA from the agarose gel: digestion of the agarose by b-agarase (Gelase, Epicentre, Madison, WI) or by
membrane dialysis. For
ligation into vector V41, the molar ratio of the vector to the insert DNA was
4:1. After 24 h incubation at 11°C, ligated DNA was transformed by
electroporation into the E.coli strain
ElectroMAX DH10B using Gibco BRL Cell Porator and Voltage Booster system (Gibco
BRL, Grand Island, NY). Recombinant transformants were selected on LB agar plate
containing 15 mg/l tetracycline, 0.5 mM IPTG and 50 mg /ml X-gal. After 24 h
incubation at 37°C, individual white colonies were picked into 384 well plates
containing LB freezing medium using Flexsys robotic workstation (Genomic
Solutions, Ann Arbor, MI) or by hand using sterile wooden toothpicks. The plates
were incubated at 37°C for 14 h and stored at -80°C. The average insert size
was assayed after plasmid DNA extraction from BACs using the method of Sambrook
et al., (10) restriction digestion with NotI
to free the DNA insert from the vector and size separation by PFGE.
The fragments were stained with ethidium bromide and visualized with
ultraviolet light.
Working copies of the BAC library were produced using a Flexsys robotic
workstation and by hand using a 384 pin replicator (Nunc, Naperville, IL).
The replicator was used to spot 6912 clones of the library onto Hybond N+
filters (Amersham-Pharmacia, Piscataway, N.J.). The inoculated filters were placed on plates containing LB
agar and 25 mg tetracycline and incubated overnight at 37°C.
The filters were prepared for hybridization with random hexamer 32P-labeled
DNA probes of oligomer repeat primer (ATATATATATATATATATAT) (10).
Results
The isolation of DNA using the method of Zhang (13) resulted in
concentrated high molecular weight DNA. Optimization
of restriction enzyme concentration indicating the use of very low
concentrations of enzyme (0.1-1 units per 1 mg of DNA) to achieve the desired partial digest of the
DNA. Both methods of the isolation
of size-selected DNA fragments resulted in high numbers of fragments for
ligating into V41. The first method
of b-agarase
enzymatic digestion of the agarose was used for the first 20,000 packaged BAC
clones and the method using membrane dialysis was used for the next 30,000
cloned fragments. Partial DNA
digests using HindIII produced high
quality and quantity transformable ligations than did restriction digests and
ligations than with BamHI digests
(data not shown). Therefore, we
used only HindIII partial digests of
the HMW DNA to complete the 50,000 BACs for the one genome equivalent coverage
of pea. We tested using one size
selection PFGE of the HMW DNA before ligation.
The number of transformants was very high (20,000 per ml ligated DNA/ml of
competent cells). However, the
average insert size (£60 kb) would increase the number of clones
significantly to achieve good coverage of the pea genome.
We also ran the two size-selection PFGE conditions on one gel.
This also resulted in an undesirable decrease in average insert size. Therefore, two size-selection gels were used to prepare all
the DNA for ligation into V41.
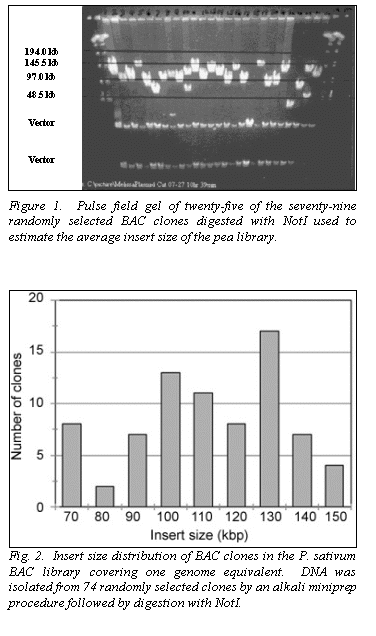 Using the parameters described, we packaged 50,000 large-insert BACs from
the pea genome. BAC insert DNA was
isolated by miniprep, digested with the restriction enzyme NotI, electrophoresed using the PFGE, and visualized with ethidium
bromide and ultraviolet light to estimate the average insert size (Fig. 1).
The distribution of seventy-nine BACs indicate an average size of 110 kb
(Fig. 2). The library has been duplicated to provide a working copy for
preparation of filters for hybridization. The
oligomeric repeat primer hybridized to the BAC inserts indicating the
microsatellite strategy may be useful in pea.
Using the parameters described, we packaged 50,000 large-insert BACs from
the pea genome. BAC insert DNA was
isolated by miniprep, digested with the restriction enzyme NotI, electrophoresed using the PFGE, and visualized with ethidium
bromide and ultraviolet light to estimate the average insert size (Fig. 1).
The distribution of seventy-nine BACs indicate an average size of 110 kb
(Fig. 2). The library has been duplicated to provide a working copy for
preparation of filters for hybridization. The
oligomeric repeat primer hybridized to the BAC inserts indicating the
microsatellite strategy may be useful in pea.
Discussion
A
BAC library of Pisum sativum,
representing one genome equivalent, has been constructed of partially
HindIII-digested DNA from the
pea germplasm line PI 269818 from the NPGS in the binary vector pCLD04541.
The library contains 50,000 BAC clones with an average insert size
of 110 kb. The Pisum
genome is estimated to be 3947 to 4397 Mbp/1C as determined by flow cytometry
(3) so we estimate this library represents one haploid genome equivalent.
In addition, we have packaged 20,000 mini-BACs (£60
kbp) which may be useful in filling gaps in a 5- to 6-fold coverage library. The
final library of PI 269818 of approximately 250,000 clones will represent five
to six-fold haploid genome equivalents, depending on the average insert size of
the BAC clones. Map-based cloning
using large-insert libraries has been useful for identifying candidate plant
resistance genes (2). The binary
vector, V41, allows for the direct transformation of candidate gene BACs into
plants to verify the phenotypic effect of the putative function of the gene.
The BACs will be sequenced for targeted microsatellite development,
recently demonstrated in soybean (4, 9). Markers
linked to traits of interest are used as probes to identified
BACs which will be sequenced to discover microsatellites around the
marker loci. The sequence data will
be used to first identify the simple sequence repeats (SSR) and second, design
primers around the SSR to convert the information into a co-dominant PCR marker.
These markers, called sequence-tagged microsatellites (STMS) will be
useful for genetic mapping, marker-assisted selection for genes underlying
resistance, and fingerprinting and assessing genetic diversity in pea germplasm
resources. The systematic sharing
of the BAC sequence information and published microsatellites markers will
potentially speed the discovery of positive allele/loci available in the Western
Regional Plant Introduction Station Pisum
sativum germplasm collection. DNA
has been isolated from most of the accessions in the NPGS and is available for
STMS analyses. Now that the 50,000 large insert pea BAC clones are packaged,
mapped pea probes will be used to identify BACs containing the markers to
verify that the library is representative of the genome (6).
The library will be available from the corresponding author (coynec@wsu.edu)
for collaborative research with pea geneticists.
Acknowledgment: The authors would like to
gratefully acknowledge the technical assistance of Jeff Shultz, Leon Razai and
Eowyn Corcoran Elan. This research
was funded in part by grants from the Northwest Agriculture Research Foundation
to Coyne and Inglis, USDA-CREES Special Grant program for Cool Season Food
Legumes to Inglis, Coyne and McPhee and contribution form Southern Illinois
University at Carbondale (Meksem and Lightfoot).
1.
Abelson, P.H. and Hines P.J. 1999.
Science 285:3 67-389.
2.
Arondel, V., Lemieux B., Hwang, I, Gibson, S., Goodman, H.M., Somerville,
C.R. 1992.
Science 258: 1353-1355.
3.
Arumuganathan, K. and Earle, E.D. 1991.
Plant Mol. Biol. Rep. 9: 208-219.
4.
Cregan, P.B., Mudge, J., Fickus, E.W., Marek, L.F., Danesh, D., Denny,
R., Shoemaker, R.C., Matthews, B.F., Jarvik, T. and Young N.D.
1999. Theor. Appl. Genet.
98: 919-928.
5.
Coyne, C.J., Meksem, K., Keller, K., Martin, R. and Lightfoot, D.A.
1999. Agronomy Abstracts pp.
166.
6.
Hall, K.J., Parker, J.S. and Ellis, T.H.N.
1997. Genome 40: 744-745.
7.
Hampton, R.O. 1986.
Pisum Genetics Newsletter 18: 22-25.
8.
Jones, J.D.G., Shlumukov, L., Carland, F., English, J., Scofield, S.R.,
Bishop G.J. and Harrison K. 1992.
Transgenic Res 1: 285-297.
9.
Meksem, K., Zobrist, K., Ruben, E., Hyten, D., Quanzhou, T. Zhang, H-B.
and Lightfoot, D.A. 2000.
Theor. Appl. Genet. (in press).
10. Sambrook, J.,
Fritsch, E.F. and Maniatis, T. 1989.
Cold Spring Harbor Laboratory. Cold Spring Harbor, NY.
11. Tao-Quanzhou,
and Zhang, H-B. 1998. Nucleic
Acids Research 26: 4901-4909.
12. Tanksley,
S.D. and McCouch, S.R. 1997.
Science 277: 1063-1066.
13. Zhang,H-B.,
Zhao, X.P., Ding XD, Paterson, A.H. and Wing, R.A. 1995. Plant J 7:
175-184.
 Using the parameters described, we packaged 50,000 large-insert BACs from
the pea genome. BAC insert DNA was
isolated by miniprep, digested with the restriction enzyme NotI, electrophoresed using the PFGE, and visualized with ethidium
bromide and ultraviolet light to estimate the average insert size (Fig. 1).
The distribution of seventy-nine BACs indicate an average size of 110 kb
(Fig. 2). The library has been duplicated to provide a working copy for
preparation of filters for hybridization. The
oligomeric repeat primer hybridized to the BAC inserts indicating the
microsatellite strategy may be useful in pea.
Using the parameters described, we packaged 50,000 large-insert BACs from
the pea genome. BAC insert DNA was
isolated by miniprep, digested with the restriction enzyme NotI, electrophoresed using the PFGE, and visualized with ethidium
bromide and ultraviolet light to estimate the average insert size (Fig. 1).
The distribution of seventy-nine BACs indicate an average size of 110 kb
(Fig. 2). The library has been duplicated to provide a working copy for
preparation of filters for hybridization. The
oligomeric repeat primer hybridized to the BAC inserts indicating the
microsatellite strategy may be useful in pea.