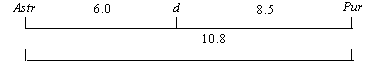
Linkage relationships of loci d, Pur and Astr
†
Gorel, F.L., Berdnikov, V.A.
†††††††††††††††††††††††††Institute of Cytology and Genetics
Rozov, S.M. and Kosterin,
O.E. †††††††††††††††††††††††††††††††Novosibirsk 630090, Russia
The region of the pea genome containing locus d (anthocyanin ring in the leaf axil) also includes several other genes (Pur, And, Ans, rup, sru) responsible for anthocyanin pigmentation in various parts of the plant (see 4). Recently, it was shown in our laboratory that the gene Astr, which determines the anthocyanin stripes on the pod wall in many strains of P. fulvum, is also linked to the gene d [1]. Also in the vicinity of d there is another locus, Pur, affecting anthocyanin distribution in the epidermis of the pod wall and funicles. It is possible that Astr might be an allele of gene Pur specific to P. fulvum. To examine this question we used the line Asteroid carrying dominant alleles of genes d, Pur and Astr. Synthesis of this line was as follows: among F2 plants from the cross cultivar Alpha (a, D, astr, pur) • P. fulvum (VIR-6070), one vigorous and fertile plant with phenotype Astr was chosen and its descendant, after selfing, was crossed with line WL-1255 (A, D, astr, Pur). A plant with phenotype D, Astr, Pur was chosen from the F2 and crossed again with line WL-1255. The presence of Pur was detected by examining the colour of the funicles. Gene Astr is reliably determined both in the field and in the greenhouse and can be used as a reliable genetic marker. After three generations of selfing, the pure breeding line Asteroid was established and crossed with WL-1514 (astr, d, pur). The F2 data (Table 1) were analysed using S.M. Rozovís program CROS. They generate the following linkage map.
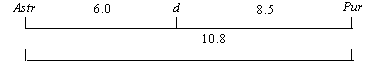
These results show that Astr and Pur are separate loci separated by a substantial map distance.
Assignment of gene d to any linkage group still remains uncertain. Recently it was shown that the breakpoint of the Twt-translocation is linked both to the gene d and to markers of linkage group IA [2, 3]. The cytological analysis of crosses with the set of standard translocations indicates that the other component of the Twt-translocation must be the chromosome corresponding to linkage group VI.
We crossed Asteroid with the Twt-translocation line (Twt, D, pur, astr) and the F2 data are given in Table 2. A heterozygous state for Twt was detected by 50% sterility of pollen. The recombination relationships obtained permit us to construct a map, where the breakpoint of the Twt-translocation is situated closer to locus Astr than locus Pur.
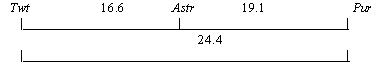
The question arises as to which gene of P. sativum is allelic to Astr of P. fulvum. If locus Astr is placed on the linkage map 6 cM from d in the opposite direction to Pur (see diagram), it maps in the vicinity of gene rup which manifests in numerous anthocyanin dots and short stripes on the pod wall. We have seen a similar phenotype in some strains of wild pea (lines L-100 and L-101 from N. Weedenís collection) formally belonging to P. sativum ssp humile but sharing some traits typical of P. fulvum. One can suppose that rup is a recessive allele of Astr and we plan to check this hypothesis.
Table 1. Joint segregation data in the F2 of cross WL-1514 (astr, d, pur) • Asteroid (Astr, D, Pur).
|
Gene pair |
Number or progeny with phenotype1 |
Joint |
RCV |
SE |
||||
| † | † |
A/B |
A/b |
a/B |
a/b |
Chi-sq |
† | † |
|
Astr |
d |
68 |
2 |
3 |
17 |
63.0*** |
6.0 |
2.6 |
|
Astr |
Pur |
63 |
6 |
3 |
17 |
47.1*** |
10.8 |
3.5 |
|
d |
Pur |
65 |
6 |
1 |
17 |
55.4*** |
8.5 |
3.1 |
1 A/a first gene; B/b second gene. ***P < 0.0001.
Table 2. Joint segregation data in the F2 of cross Twt • Asteroid.
|
Gene pair |
Phase |
Number of progeny with phenotype1 |
Joint |
RCV |
SE |
||||||
| † | † | † |
A/B |
A/h |
Ab |
a/B |
a/h |
a/b |
Chi-sq |
† | † |
|
Astr |
Twt |
C |
23 |
56 |
4 |
2 |
10 |
18 |
43.3*** |
16.6 |
3.8 |
|
Pur |
Twt |
C |
23 |
40 |
2 |
2 |
24 |
20 |
34.1*** |
24.4 |
4.6 |
|
Astr |
Pur |
C |
63 |
† |
20 |
2 |
† |
26 |
40.8*** |
19.2 |
4.2 |
1 A/a first gene; B/b second gene; h, heterozygous for Twt. When both genes are dominant, the capital letter stands for the dominant allele. When the second gene is codominant, the capital A stands for the dominant allele of the first gene and capital B for an allele of the second gene which is in coupling with A. ***P < 0.0001.
†
Acknowledgement: This work was partially supported by the Russian State Program for Fundamental Research.
_________
1. Bogdanova, V.S., Trusov, Y.A., Kosterin, O.E. 1995. Pisum Genet. 27:1-4.
2. Gorel, F.L., Temnykh, S.V., Lebedeva, I.P., Berdnikov, V.A. 1992. Pisum Genet. 24:48-51.
3. Temnykh, S.V., Gorel, F.L., Berdnikov, V.A., Weeden, N.F. 1995. Pisum Genet. 27:23-25.
4. Weeden, N.F., Swiecicki, W.K., Ambrose, M., Timmerman, G.M. 1993. Pisum Genet. 25:4 and cover.
* * * * *