|
Pisum Genetics |
Volume 26 |
1994 |
Research Reports |
pages 41-43 |
Location of the Lv gene in pea linkage group VI
|
Weller, J.L. and Murfet, I.C. |
Department of Plant Science, University of Tasmania Hobart , Tasmania 7001, Australia |
Mutant lv plants are characterised by elongated internodes when grown under red or white light but are indistinguishable from wild type Lv plants in darkness or under far-red light (4, 11). Mutant plants also lack a normal elongation response to end-of-day far-red light and to a low red : far-red ratio (10) and are earlier flowering under short (non-inductive) photoperiods (11). Four recessive lv alleles have been identified from the mutant lines NEU3, R83, Wt10895 and L80m (5, 9), with each conferring a similar phenotype (9, 11). Because expression of the Lv-lv difference is restricted to certain light conditions, the lv mutants can be termed photomorphogenic. The syndrome of photomorphogenic abnormalities seen in the lv mutants is indicative of a reduction in the function of phytochrome B (phyB), one of several related photoreceptor proteins which play a major role in the control of plant development by light. Recent results have shown that lines R83, Wt10895 and L80m are all deficient in the phyB apoprotein while the NEU3 mutant has normal levels of phyB (11) suggesting that Lv may be a structural gene for phy B.
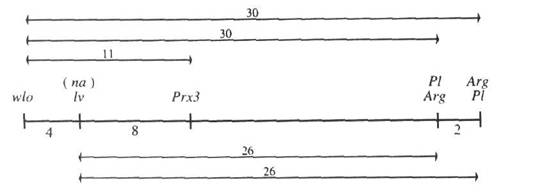
We report here data showing that the lv locus is in linkage group VI between wlo and Prx3, and close to na (within 2 cM).
F2 segregation data for lv and group VI primary markers wlo, na, Prx3, Arg and Pl [see mapping guidelines (8)] were obtained from three crosses as detailed in Table 1. Parental marker lines 111 (A875-55-0) and 224 (A783-161) come from the Marx collection, line 107 is a selection from cv Torsdag, and the lv allele in line 232– is derived from mutant line NEU3. Further details of the lines used are given in previous papers (9, 10).
Identification of the Lv - lv segregation was facilitated by growing the plants for the first 10 days in a growth chamber at 20°C under continuous white light (150 (mmol m-2s-1 at pot top) supplied by 40W cool white fluorescent tubes. The plants were then transferred to the glasshouse and grown to maturity under an 18 h photoperiod. All crosses were of normal fertility. Data were analysed using the programs LINKAGE-1 (6) and CROS (S.M. Rozov).
All individual segregations in Table 1 are in accordance with expectation (P>0.05). The joint segregation data reveal strong linkage of lv with na (<2 cM), wlo (4 cM) and Prx3 (8 cM) and moderate linkage with Arg (26 cM) and Pl (26 cM) with P<0.000001 and <0.0001, respectively.
Our
data for wlo - Prx3 generate a map distance about one third that shown in
the latest map (7). However, our data for wlo - Pl and wlo -
Arg are consistent with the latest map and values obtained from very
large data sets by Lamprecht (1) and Marx (2, 3). Based on a sample of 2797
plants, Lamprecht reported a recombination fraction of 31.6 ± 1.1% for wlo and
Pl. Marx' data indicate a similar value for wlo and
Table 1. F2 segregation data for lv and linkage group VI markers.
|
Loci |
Crossa |
Phenotypeb |
Total |
Chi-squared |
Linkage |
Recomb. |
SE |
||||||||
|
Locus 1 |
Locus 2 |
Joint |
|||||||||||||
|
|
|
|
DD |
DR |
RD |
RR |
|
|
|
|
|
|
|
|
|
|
Lv |
Wlo |
1 |
99 |
2 |
3 |
24 |
|
|
128 |
1.04 |
1.50 |
99.41 |
<0.0001 |
4.2 |
1.8 |
|
Lv |
Arg |
1 |
87 |
14 |
13 |
14 |
|
|
128 |
1.04 |
0.67 |
17.99 |
<0.0001 |
26.2 |
4.7 |
|
Lv |
Pl |
1 |
87 |
14 |
13 |
14 |
|
|
128 |
1.04 |
0.67 |
17.99 |
<0.0001 |
26.2 |
4.7 |
|
Wlo |
Arg |
1 |
86 |
16 |
14 |
12 |
|
|
128 |
1.50 |
0.67 |
11.25 |
<0.001 |
30.2 |
5.0 |
|
Arg |
Pl |
1 |
99 |
1 |
1 |
27 |
|
|
128 |
0.67 |
0.67 |
116.56 |
<0.0001 |
1.7 |
1.1 |
|
Wlo |
Pl |
1 |
86 |
16 |
14 |
12 |
|
|
128 |
1.50 |
0.67 |
11.25 |
<0.001 |
30.2 |
5.0 |
|
Lv |
Wlo |
2 |
45 |
30 |
24 |
0 |
|
|
99 |
0.03 |
1.48 |
13.77 |
<0.001 |
|
|
|
Lv |
Na |
3 |
130 |
3 |
2 |
46 |
|
|
181 |
0.22 |
0.41 |
156.44 |
<0.0001 |
2.7 |
1.2 |
|
|
|
|
DF |
DH |
DS |
RF |
RH |
RS |
|
|
|
|
|
|
|
|
Lv |
Prx3 |
2 |
3 |
32 |
25 |
19 |
3 |
0 |
82 |
0.15 |
1.98 |
54.83 |
<0.0001 |
7.6 |
3.0 |
|
Wlo |
Prx3 |
2 |
22 |
32 |
6 |
0 |
3 |
19 |
82 |
0.15 |
1.98 |
44.80 |
<0.0001 |
11.0 |
3.6 |
aCross: 1) line 80m (lv wlo arg pl) x line 224 ( Lv Wlo Arg Pl)
2) line 232– (lv Wlo Prx3F) x line 111 (Lv wlo Prx3S)
3) line 107 (Lv Na) x lv na segregate from cross NEU3 (lv Na) x L81 (Lv na)
bD = dominant, R = recessive, F = homozygous fast, H = heterozygous, and S = homozygous slow. The first named locus is shown first.
The data in Table 1 generate the following map:

In summary, these results obtained from three different crosses and using two different lv alleles are consistent and they provide convincing evidence that lv is located in linkage group VI between wlo and Prx3 and close to na. We have planned a 5-point coupling phase cross involving standard line JI1794 and markers wlo, lv, Gty, Prx3 and Pl to further examine distances in this section of group VI and other crosses to determine the position of na.
Acknowledgments. We thank the Australian Research Council for financial support and Dr S.M. Rozov for use of the program CROS.
Lamprecht, H. 1961. Agri Hort. Genet. 19:360-401.
Marx, G.A. 1981. Pisum Newsl. 13:35-37.
Marx, G.A. 1982. Pisum Newsl. 14:50-52.
Nagatani, A., Reid, J.B., Ross, J.J., Dunnewijk, A. and
Furuya, M. 1990. J. Plant
Physiol. 135:667-674.
Reid, J.B. and Ross, J.J. 1988. Physiol. Plant. 72:595-604.
Suiter, K.A., Wendel, J.F. and Case, J.S. 1983. J. Hered. 74:203-204.
Weeden, N.F., Ambrose, M. and Swiecicki, W.K. 1991. Pisum Genet. 23 cover.
Weeden, N.F., Swiecicki, W.K., Ambrose, M. and
Timmerman, G.M. 1993. Pisum
Genet. 25:4 and cover.
Weeden, N.F., Swiecicki, W.K., Timmerman, G.M. and
Ambrose, M. 1993. Pisum
Genet. 25:13-14.
Weller, J.L., Murfet, I.C. and Reid, J.B. 1992. Pisum Genet. 24:86-89.
Weller, J.L. and Reid, J.B. 1993. Planta 189:15-23.
Weller, J.L., Nagatani, A., Kendrick, R.E., Murfet, I.C.
and Reid, J.B. 1995. Plant
Physiol. (in press).