|
Pisum Genetics |
Volume 26 |
1994 |
Review |
pages 1-5 |
Recent advances in gene transfer to peas
|
Schroeder, H.E., Gollasch, S., Tabe L.M. and Higgins, T.J.V. |
CSIRO Division of Plant Industry GPO Box 1600 , Canberra , ACT 2601, Australia |
The development of genetic engineering procedures for the introduction of foreign genes into peas has proved a difficult task. However, four years ago Puonti-Kaerlas (6) produced the first transgenic pea plants. Stable transformation was achieved by using resistance to the antibiotic hygromycin as the selectable marker. On further analysis it was found that all transgenic plants and progeny were aberrant types. Using kanamycin selection and B-glucuronidase as a reporter gene, Davies et al. (1) produced four transgenic pea lines. The method proved to be difficult to reproduce. Davies and Mullineaux (2) concluded that the system needed further development.
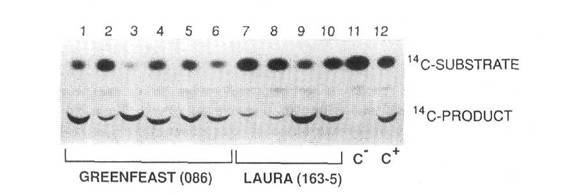
We have reported the development of a routine reliable transformation and regeneration system for peas (9). This system was established using Agrobacterium-mediated gene transfer to introduce two chimeric genes, an antibiotic resistance gene (nptII) and a herbicide resistance gene (bar) into two cultivars of pea, Greenfeast and Rondo. The expression of the nptII and bar genes in primary transgenics and first generation progeny was confirmed by enzyme assays. It was found that the bar gene was an efficient selectable marker in the tissue culture phase of pea transformation. This gene confers resistance to phosphinothricin (PPT), the active ingredient in the non-selective herbicide Basta. The bar gene encodes the enzyme phosphinothricin acetyl transferase (PAT) which catalyses the conversion of PPT to a non-toxic acetylated product (Fig. 1). The bar gene is also a screenable marker with great potential for use in conventional breeding when foreign genes from transgenic plants are transferred to existing commercial cultivars. Either painting of individual leaves or spraying of plants with the herbicide will indicate expression of the bar gene (9) and because of linkage to the other genes in the introduced construct, will serve as a scoreable phenotypic marker. Although gene transfer into peas in our laboratory was established using the garden pea cultivars Greenfeast and Rondo, the procedures have now been extended to the transfer of useful genes into Australian field pea cultivars Dundale and Laura.
As a result of discussions with pea breeders, pea growers, and pea marketers, the initial aims of our pea crop improvement program are to introduce three new traits, namely, resistance to the insect pest, pea weevil (Bruchus pisorum), tolerance to the herbicide Basta, and improved nutritional quality of pea seed proteins.
Bean a-amylase inhibitor confers resistance to Bruchid weevils attacking stored grain
The fully matured, stored seeds of peas and other grain legumes such as chickpeas, cowpeas and Azuki beans are susceptible to infestation by seed-feeding bruchids (Callosobruchus sp.). The seeds of another grain legume, the common bean (Phaseolus vulgaris), are resistant to these seedfeeding bruchids because of the presence of a seed protein with high insecticidal activity, the a-amylase inhibitor protein (aAI). Studies with artificial diets (3, 4) showed that the development of two seed-feeding beetles, the cowpea weevil (Callosobruchus maculatus) and the Azuki bean weevil (C. chinensis), was inhibited by relatively low levels of bean aAI in the diet. This information prompted us to investigate whether transfer and expression of the bean aai gene in peas might confer protection against these pests of stored grain legumes.

Fig. 1. Expression of the bar gene in transgenic peas. Expression was measured as phosphinothricin acetyl transferase activity in the leaves of six primary regenerants of cv Greenfeast (lanes 1-6) and in four primary regenerants of cv Laura (lanes 7-10). Lane 11 contains extract from leaves of a nontransformed pea plant (C). Lane 12 contains extract from leaves of a transgenic tobacco expressing the bar gene (C).
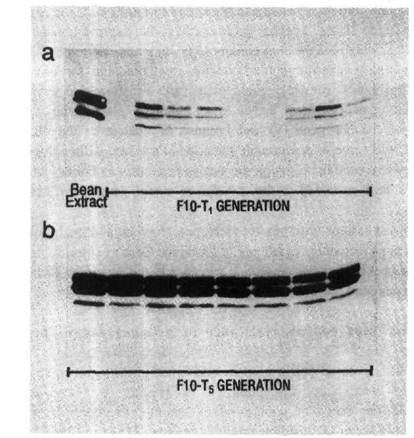
Fig. 2. The distribution of aAI protein in individual transgenic pea seeds: Western blots of aAI protein in: a) nine T1 seeds of a line showing segregation of the aai product; b) eight T5 seeds of a line showing homozygous expression of the aai gene. Bean seed extract served as a positive control for aAI protein. (Reproduced with permission from Plant Physiology)
The bean aai gene was modified by replacing its promoter with the bean phytohemagglutinin promoter and the modified aai gene was introduced into the pea cultivar Greenfeast (10). Selected seeds of the second transgenic generation (T2) of several transgenic lines were then used to test for resistance to C. maculatus and C. chinensis. To obtain T2 seeds, T1 seeds were screened by immunoblot procedures (Fig.2a) and only those seeds which tested positive for the aAI protein, were raised to produce T1 plants with T2 seeds. Due to segregation of the transgenes, the level of aAI protein in T2 seeds ranged from undetectable to 3.5% of total soluble seed protein. Transgenic pea seeds were bioassayed for bruchid resistance by infesting them with either C. chinesis or C. maculatus. Larval development times, within seed development time (WSDT), and within seed mortality (WSM) were recorded in both transgenic and untransformed control seed (10). The results indicated that low levels of aAI in pea seeds conferred virtual immunity against the Azuki bean weevil. The lowest level of aAI tested (0.15% w/w) killed all weevils. In contrast, resistance of transgenic seed to cowpea weevils was proportional to the level of aAI (Fig. 3), and complete inhibition of the cowpea weevil development was only achieved with a higher level of aAI (0.77% w/w). There was excellent correspondence between determined aAI levels and mortality and delayed development of cowpea weevil larvae as indicated by correlations between WSM and aAI content, r = 0.948, and WSDT and aAI content, r = 0.956.
Resistance to pea weevils in the field
The pea weevil (Bruchus pisorum) is the major insect pest of pea crops in Australia; it attacks the developing fruit and matures within the ripening seeds. Bioassay experiments were set up to investigate pea weevil infestation and development and to ascertain whether the presence of the aAI protein in pea seeds could protect the growing crop from insect attacks. Because of the transgenic status of the plants, bioassays had to be conducted in a biosafety glasshouse. Two pea weevil bioassays were carried out, the first with T2 seeds of five transgenic lines, the second with T5 seed from one of the transgenic lines (8).
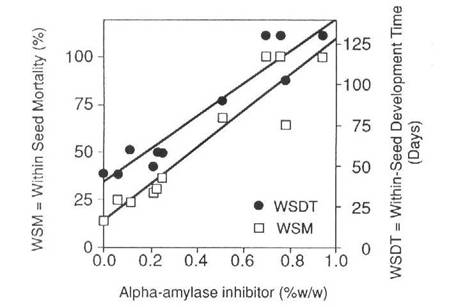
Fig. 3. Time taken to reach maturity and mortality of cowpea weevil larvae in transgenic pea seeds expressing various levels of aAI protein. (Reproduced with permission from Bio/Technology).
In the first bioassay, T1 seeds, positive for aAI protein, were used to produce Tl plants producing pods for simulated weevil infestation. Infestation was achieved by transferring pea weevil eggs to immature pods. Together with each transgenic pod, a control pod on a nontransgenic pea plant was infested at the same time. Pods were harvested at seed maturity and the testa and cotyledons of every seed were examined to ascertain the number of infested seeds and the number of emerged adult weevils. Seeds which had been scored as infested but from which no adult weevil had emerged after 140 d from the date of egg transfer were split open for more detailed examination. In the first bioassay the level of aAI protein in mature T2 seeds ranged from undetectable to over 3% of total soluble seed protein. The variation in aAI content of the seeds was attributable to two factors. Firstly, seed from several different trangenic pea lines were tested, and secondly, the inherited aai transgene was segregating in the T2 populations. In the transgenics with the higher aAI contents, adult weevils emerged only in seeds which did not contain aAI. Seeds from one transgenic line (F10) with the highest aAI content (>3% of total soluble seed protein), were used to produce T4 plants which were homozygous for the aai gene and produced uniformly high levels of aAI protein in all the seeds (Fig. 2b). Immature pods on T4 plants were used in the second bioassay. Conditions of infestation were identical to bioassay 1. The mean time to adult weevil emergence in nontransgenic control seeds was 85 d. After 140 d no adult pea weevil had emerged from any T5 transgenic seed and additional observation after 200 d indicated that total inhibition of pea weevil development had been achieved.
The seed-specific aai gene has now been stably expressed in transgenic pea seeds for six generations with no change in the level of aAI expression. Transgenic plants bearing this seed are phenotypically similar to control plants (Fig. 4 - see volume 26 cover and legend). Currently we are using genetic engineering procedures and conventional breeding techniques to transfer the bean aai gene into commercial cultivars of field peas. Crosses between the F10 T5 homozygous Greenfeast line and cvs Dun, Dundale, Bluey and Laura have been made and the aAI protein has been detected in mature hybrid seeds. Homozygotes will be identified for backcrossing to the respective cultivars. There are excellent prospects for controlling the pea weevil field pest by the introduction of the aai gene.
Improving the amino acid composition of the seed protein
The pea is a rich source of protein which is free of antinutritional factors but, in common with most other grain legumes, peas are a poor dietary source of the essential sulphur-containing amino acids methionine and cysteine. The globulins, legumin and vicilin, together with a small number of albumins constitute the bulk of the storage proteins of pea seeds. Although the relative proportions of these seed storage proteins vary in different lines of pea, the total sulphur amino acid content of all lines appears to be similar (7). Since little or no variation exists within the genus Pisum to increase the sulphur amino acids by selective breeding, there is an opportunity to use genetic engineering to rectify this situation.
A sunflower gene encoding a 2S seed albumin (SFA8), which contains 24% methionine and cysteine (5), was modified for expression in pea. Its 5' and 3' flanking regions were replaced with the corresponding regions from the pea vicilin gene. This chimeric Vc-SFA8 gene was transferred to peas and was expressed in transgenic pea seeds. The level of SFA8 protein in a number of transgenic pea lines was estimated at approximately 0.5% of total soluble seed protein. On amino acid analysis of total soluble seed protein this translated into a modest increase in Met + Cys of 17 to 22%. In the third seed generation (T3), several plants homozygous for the SFA8 gene have been selected. Western blot analyses indicate an SFA8 expression level of around 1% of total soluble seed protein, which could translate into a 30% increase in seed protein sulphur amino acid content. We will cross different transgenic pea lines homozygous for the SFA8 gene in an attempt to increase total S-amino acid levels by over 50%.
In summary, we have demonstrated the successful introduction of several foreign genes into peas. The genes are expressed in the target organs, e.g. the whole plant in case of the bar gene and the seed in the cases of the aai gene and the sunflower albumin gene. The introduced seed-specific genes have been stably expressed for a number of generations and have been transmitted to nontransgenic peas by conventional crosses. We are now targeting fungal and viral resistance, as well as resistance to another serious insect pest in the field, Helicoverpa punctigera.
Acknowledgement: This research was supported by the Grains Research and Development Corporation.
Davies, D.R., Hamilton, J. and Mullineaux, P. 1993. Plant Cell Reports 12:180-183.
Davies, D.R. and Mullineaux, P.
1993. In Peas: Genetics, Molecular Biology and
Biotechnology, Eds R. Casey and D.R. Davies, CAB International, Wallingford,
U.K.,
pp. 291-301.
Huesing, J.E., Shade, R.E.,
Chrispeels, M.J. and Murdock, L.L. 1991. Plant
Physiol. 96:993-996.
Ishimoto, M. and Kitamura, K. 1989. Appl. Entomol. Zool. 24:281-286.
Kortt, A.A., Caldwell, J.B., Lilley,
G.C. and Higgins, T.J.V. 1991 Eur. J. Biochem.
195:329-334.
Puonti-Kaerlas, J., Erickson, T. and
Engstrom, P. 1990. Theor. Appl. Genet. 80:246-
252.
Schroeder, H.E. 1982. J. Sci. Food Agri. 33:623-633.
Schroeder, H.E., Gollasch, S.,
Moore, A., Tabe, L.M., Craig, S., Hardie, D.C.,
Chrispeels, M.J., Spencer, D. and Higgins, TJ.V. 1995. Plant Physiol. (in
press).
Schroeder, H.E., Schotz, A.H.,
Wardley-Richardson, T., Spencer, D. and Higgins,
T.J.V. 1993. Plant Physiol. 101:751-757.
Shade, R.E., Schroeder, H.E., Pueyo,
J.J., Tabe, L.M., Murdock, L.L., Higgins, T.J.V.
and Chrispeels, M.J. 1994. Bio/Technology 12:793-796.