|
Pisum Genetics |
Volume 25 |
1993 |
Research Reports |
pages 64-70 |
Seed mutants in Pisum
|
Wang, T.L. and Hedley, C.L. |
John Innes Institute John Innes Centre, Colney, Norwich , NR4 7UH , UK |
Three new rugosus loci
Until recently, alleles at only two loci were known to affect embryo development in peas and their storage product composition. These loci, the rugosus loci r (17) and rb (9), cause the dry seed to be wrinkled. They also alter the amount and composition of the starch and protein and the level of lipid.
In 1987 we initiated a chemical mutagenesis programme to isolate additional wrinkled-seeded mutants. A total of twenty thousand seeds of a round-seeded line (BC1/RR; 8) were treated either with ethyl methane sulphonate (EMS) or methyl nitroso urea (NMU) and the progeny screened for wrinkled seeds. Preliminary data on the results of the programme have been published (14) and also data on the storage product composition of seeds from early generation material (15). Each mutant was assigned a SIM (Seed : induced mutant) number at this stage. These data indicated that a number of mutants had been generated encompassing a range of storage product compositions. Preliminary genetic analysis had shown that at least three new loci had been mutated though several mutants were unassigned (7, 16).
The mutants have been maintained in three ways. Firstly, they have been purified by single-seed descent, currently for 7 or 8 generations. Secondly, they have been purified by selfing of and re-selecting plants heterozygous for the mutation for 8 generations to date. From this process, therefore, pairs of near-isolines for each mutation can be selected though they will be in backgrounds which may differ from the parental line and from pair to pair. Finally, the lines are in the process of being backcrossed to the parental line (BC1/RR) to produce near-isolines all in the same background. Selections of mutants following 3 backcrosses have been made so far. The mutants segregate when the heterozygous plants are selfed, or at the F2 when the mutants are backcrossed, consistent with their being monogenic recessives (Table 1).
The initial complementation analysis was carried out using a full diallel with plants from wrinkled seeds originating from either the single seed descent or heterozygote lineages. The analysis, however, was incomplete since several crosses gave progeny whose phenotype was ambiguous with respect to the shape of the seed. A half diallel has recently been carried out to complete the data, using representatives of the five loci from the purified lines and the undefined mutants. Table 2 shows the results of the complete complementation analysis.
In accordance with the Pisum Genetics Association gene symbol committee, we have assigned the gene symbols, rug-3, rug-4, rug-5 for the three new rugosus loci.
Table 1. Segregation analysis of seed from sister plants of the third backcross of the SIM lines.
|
SIM No. |
No. of plants |
Round seed |
Wrinkled Seed |
c2(3:1) |
|
1 |
5 |
302 |
116 |
1.68 |
|
11 |
5 |
223 |
73 |
0.02 |
|
14 |
5 |
277 |
74 |
2.87 |
|
15 |
5 |
224 |
87 |
1.47 |
|
16 |
5 |
240 |
91 |
1.10 |
|
31* |
5 |
365 |
106 |
1.56 |
|
32 |
5 |
249 |
79 |
0.15 |
|
41 |
3 |
128 |
52 |
1.45 |
|
42 |
5 |
264 |
75 |
1.50 |
|
43 |
5 |
218 |
81 |
0.70 |
|
51 |
5 |
242 |
72 |
0.72 |
|
52 |
4 |
171 |
55 |
0.05 |
|
53 |
5 |
200 |
73 |
0.44 |
|
54 |
4 |
167 |
51 |
0.30 |
|
55 |
5 |
194 |
82 |
3.27 |
|
56 |
5 |
229 |
66 |
1.09 |
|
57 |
4 |
178 |
55 |
0.24 |
|
58 |
3 |
163 |
41 |
2.61 |
|
59 |
5 |
248 |
66 |
2.65 |
|
61 |
5 |
255 |
80 |
0.22 |
|
71 |
4 |
190 |
39 |
7.56 |
|
81 |
5 |
234 |
78 |
0.00 |
|
91 |
5 |
207 |
69 |
0.00 |
|
101 |
5 |
209 |
63 |
0.49 |
|
102 |
5 |
256 |
93 |
0.51 |
|
103 |
5 |
220 |
62 |
1.37 |
|
103W |
5 |
200 |
56 |
1.33 |
|
201 |
5 |
205 |
87 |
3.58 |
*Seed from a selfed heterozygote as the mutant does not survive.
** P < 0.01 but seed of the selfed heterozygote (8th selection) gave 313 round : 100 wrinkled seeds (c2 = 0.16).
Table 2. Complementation groups of the rugosus mutants. The numbers indicate the original SIM numbers. Complementation groups 1 and 2 correspond to loci r and rb, respectively.
|
1 |
2 |
3 |
4 |
5 |
|
53 |
14 |
1 |
11 |
51 |
|
54 |
15 |
32 |
91 |
52 |
|
55 |
16 |
41 |
201 |
81 |
|
56 |
101 |
42 |
|
|
|
57 |
102 |
43 |
|
|
|
58 |
103 |
|
|
|
|
59 |
103W |
|
|
|
|
61 |
|
|
|
|
|
71 |
|
|
|
|
The rug symbol was chosen in order to use the 3-character nomenclature recommended. The various mutant alleles have been identified by superscript letters as shown in Table 3. The superscripts a and b have not been used for r or rb alleles in order to avoid possible confusion with the symbols ra and rb which, as stated by Blixt (2), were assigned incorrectly as the base symbol for these two loci by Kooistra (9). Examples of seeds from a line at each locus are given in Fig. 1.
Alleles at the r locus have been analysed in detail. There is some uncertainty over SIM55 since it has characteristics which make it very similar to the original rr line (WL200) in that it produces a larger transcript, indicative of an insertion of a DNA sequence (1). It may represent, therefore, a rr seed that was included by mistake in the mutagenesis of the original round seeds. SIM 56 produces neither protein nor transcript and is thus a null mutant (M. MacLeod, pers. com.).
The phenotypes caused by the new alleles at the rb locus all appear to be similar or milder (i.e. more starch, weaker wrinkling of the seed) than the rb phenotype, but have not been investigated in detail. Two lines of note, however, are SIM103 and 103W. These lines originated from a single line from which two lines segregated (one with more severe wrinkling than the other). Subsequent analysis showed that the two bred true on the basis of wrinkling and storage product content and that both mutants were alleles at the rb locus.
The rug-3 locus mutants have characteristics similar to mutants of both Nicotiana and Arabidopsis (3, 5) in that mutations at the locus dramatically decrease the starch content of the seeds to as little as one hundredth of that in the wild-type, and also decrease the content in the leaves. The Nicotiana sylvestris mutant, NS458, has a modified plastidial phosphoglucomutase while in Arabidopsis, there are mutants which have lower phosphoglucomutase activity (mutant TC7, pgmPpgmP) and those that lack ADP glucose pyrophosphorylase activity (mutant TL25, adg1adg1; 10). These mutants were isolated by screening leaves for a lack of starch. In the leaves of rug-3 rug-3 plants there is no blue colouration produced when they are treated with iodine stain following chlorophyll removal, the leaves remaining yellow.

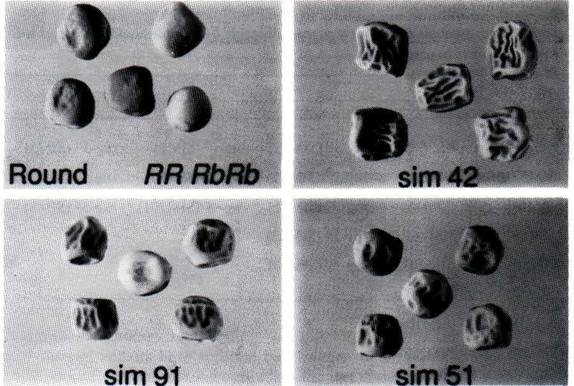
Fig. 1. Phenotypes of pea seeds. Round-seeded line (BC1/RR; 8); SIM42 rug-3rug-3; SIM91 rug-4rug-4; SIM51 rug-5rug-5.
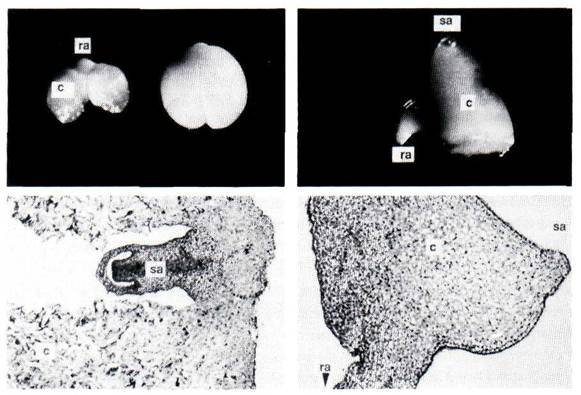
Fig. 2. Examples of embryo morphology mutants. Top row - morphology of isolated embryos: left - mutant E1735 with wild-type; right - E4650. Bottom row - wax embedded sections of: left - mutant E1735; right - mutant E4650. c - cotyledon; ra-root axis; sa - shoot axis.
Table 3. Gene symbols for the new rugosus mutants by complementation group with the type line shown in parentheses.
|
1 |
2 |
3 |
4 |
5 |
|
r (WL200) |
rb (WL1685) |
rug-3a (SIM1) |
rug-4a (SIM11) |
rug-5a (SIM51) |
|
rc (SIM53) |
rbc (SIM14) |
rug-3b (SIM32) |
rug-4b (SIM91) |
rug-5b (SIM52) |
|
rd (SIM54) |
rbd (SIM15) |
rug-3c (SIM41) |
rug-4c (SIM201) |
rug-3c (SIM81) |
|
re (SIM55) |
rbe (SIM16) |
rug-3d (SIM42) |
|
|
|
rf (SIM56) |
rbf (SIM101) |
rug-3e (SIM43) |
|
|
|
rg (SIM57) |
rbg (SIM102) |
|
|
|
|
rh (SIM58) |
rbh (SIM 103) |
|
|
|
|
ri (SIM59) |
rbi (SIM103W) |
|
|
|
|
rj (SIM61) |
|
|
|
|
|
rk (SIM71) |
|
|
|
|
The rug-4 and rug-5 loci have not been studied at all as yet, other than the analysis of their starch and amylose contents. Mutations at both loci result in a decreased starch content, rug-4 less so than rug-5 which is similar to both the r and rb loci. The amylose content (as a percentage of the starch) of rug-4 mutants is similar to that of the r and rb mutants, though the amylose content of rug-5 mutants is about 30% higher than that of the wild-type (full details of the starch contents of all the mutants will be published elsewhere).
Other wrinkled-seeded mutants have also been isolated from the programme; SIM31, which was identified in the original screen, deserves a mention. The seed is wrinkled but noticeably smaller than other wrinkled types and it is khaki in contrast to the buff to pale green of the others. It also has a lower starch content than its round-seeded isoline. It germinates well and as fast as its round-seeded isoline but, in contrast to all the other mutants, the plant is pale green and dies after reaching a few inches in height and developing only a few nodes. No genetic analysis has been performed on this line but phenotypically it is distinct from the other rug loci. In addition, after re-screening some of the later populations derived from the original mutation programme, a few additional mutants with altered seed shape have been isolated. These are currently being used to generate isogenic lines.
[Any researcher wishing to collaborate in the use of these rugosus lines should contact the authors (Tel: +44 603 52571, Fax: +44 603 56844, Email: Wang@UK.AC.AFRC or HedleyC@UK.AC.AFRC)].
Table 4. Segregation analysis of embryo phenotypes of progeny from selfed heterozygote sisters (6 th selection) of embryo morphology mutants.
|
Mutant No. |
Wild-type |
Mutant |
c2(3:l) |
|
E1137 |
202 |
77 |
1.01 |
|
E1450 |
183 |
56 |
0.31 |
|
E1735 |
186 |
77 |
2.57 |
|
E1881 |
178 |
67 |
0.72 |
|
E2748 |
170 |
61 |
0.24 |
|
E4650 |
85 |
30 |
0.07 |
|
E6101 |
176 |
54 |
0.28 |
Embryo morphology mutants.
Several authors have reported the isolation of mutants affected in embryo development. In particular, there are a large number of such mutants in maize (13) and Arabidopsis. In the latter, there exist both embryo lethal mutants (12) and ones affected in pattern formation (11). We are particularly interested in the cellular development of pea embryos and how this development relates to storage product deposition (4, 6). We have taken, therefore, a genetic approach to disrupt the development of the embryo in the hope of gaining an insight into the processes that take place there. Hence, the same mutagenised seed population used for the isolation of the new rugosus mutants was employed for the isolation of mutants with defective embryos. In this instance, screening was performed by opening pods on plants when the seeds were immature and still contained liquid endosperm, and observing for segregation of embryos with defects. Initially, 30 lines were isolated, 9 of which it proved possible to maintain subsequently by the selling of the heterozygotes - E1137, E1450, E1624, E1735, E1881, E2391, E2748, E4650 and E6101. At each generation, 3 pods were opened and the embryos scored for segregation so that plants heterozygous for the mutation can be identified.
Of the 9 lines, several have similar phenotypes. E1137, E1881 and E2391 all appear as if they have single or possibly fused cotyledons. E4650 is similar but, in addition, possibly has disturbed axes (Fig. 2). E1450 and E6101 are both blocked in development and form small embryos with bulbous cotyledons, apparently arrested at the late heart shaped stage. When the seeds of E2748 are cut open, the mutants are clearly recognised by being dark green and seemingly adhering to the testa. In section, these appear to be defective in the outer cell layers of the embryo which produces a close association between embryo and testa. E1735 has a gross abnormality in the development of its cotyledon cells (Fig. 2). An equivalent to this latter phenotype does not exist in other species such as maize or Arabidopsis (pers. corns, from Bill Sheridan and Gerd Jürgens) and E1735 is thus a unique embryo mutant.
The last mutant, E1624, remains unfathomable at present. The line segregates into several phenotypes which are not easily scored, an almost normal embryo with cotyledons slightly displaced; an abnormal embryo which is displaced 180° in two dimensions within the testa; the same two phenotypes but with an apparently vestigial root. The abnormal embryo without a root germinates to produce a shoot only which, when grafted onto a normal plant, will give progeny containing all phenotypes.
Analysis of segregating populations of the lines indicated monogenic recessive inheritance of each mutant (Table 4). E1624 is excluded from this list for obvious reasons and E2391 because it is still under investigation. Problems associated with the lethal nature of some of the mutants has prevented us establishing whether any are allelic. In recent generations, however, more material has become available and we have been able to obtain some more of the lines as pure breeding. We hope, therefore, to carry out some genetic analyses in the near future. Until then we have not assigned any gene symbols to these new mutants.
Acknowledgements. We thank our colleagues Lorraine Barber, Chris Harrison, Alan Jones, Sue Johnson, Chun-Ming Liu, James Lloyd and Calum MacLeod for discussions and providing some of the data cited above. This work was supported via a grant-in-aid to the John Innes Institute from the Agricultural and Food Research Council. We also wish to acknowledge support from the Ministry of Agriculture, Fisheries and Food and AGC Cambridge, for LB from the EC ECLAIR programme, for CH from PBIC Cambridge, and for JL from Maribo Seeds, Denmark.
Bhattacharyya, M.K., Smith, A.M.,
Ellis, T.H.N., Hedley, C.L. and Martin, C.R.
1990. Cell 60:115-122.
Blixt, S. 1977. PNL 9 (supplement).
Caspar, T., Huber, S.C. and Somerville, C. 1985. Plant Physiol. 79:11-17.
Corke, F.M.K., Hedley, C.L. and Wang, T.L. 1990. Protoplasma 155:127-135.
Hanson, K.R. and McHale, R.A. 1988. Plant Physiol. 88:838-844.
Hauxwell,
A.J., Corke, F.M.K., Hedley, C.L. and Wang, T.L. 1990.
Development 110:283-289.
Hedley,
C.L., MacLeod, M., Johnson, S., Jones, A., Barber, L. and Wang, T.L.
1992. In Proceedings of the 1 st European Conference on Grain Legumes, Angers,
1992, AEP, Paris, pp. 167-168.
Hedley,
C.L., Smith, C.M., Ambrose, M.J., Cook, S.K. and Wang, T.L. 1986.
Ann. Bot. 58:371-379.
Kooistra, E. 1962. Euphytica 11:357-373.
Lin,
T-P., Caspar, T., Somerville, C. and Preiss, J. 1988. Plant Physiol.
86:1131-1135.
Mayer,
U., Ruiz, R.A.T., Berleth, T., Misera, S. and Jürgens, G. 1991. Nature
353:402-407.
Meinke, D.W. 1991. The Plant Cell 3:857-866.
Sheridan, W.F. and Clark, J.K. 1993. The Plant J. 3:347-358.
Wang,
T.L., Hadavizideh, A., Harwood, A., Welham, T.J., Harwood, W.,
Faulks, R. and Hedley, C.L. 1990. Plant Breeding 105:311-320.
Wang, T.L. and Hedley, C.L. 1991. Seed Science Research 1:3-14.
Wang,
T., MacLeod, M., Johnson, S., Jones, A., Barber, L., Martin, C. and
Hedley, C. 1993. In Basic and Applied Aspects of Seed Biology,
Proceedings of
the IV International Workshop on Seeds, Angers, Eds D. Come and F. Corbineau,
Vol. 1, ASFIS, Paris, pp. 153-159.
White, O.E. 1917. Proc. Amer. Phil. Soc. 56:487-588.