|
Pisum Genetics |
Volume 25 |
1993 |
Research Reports |
pages 40-42 |
Ramosus loci rms-3 and rms-4 are in pea linkage groups 1 and 7, respectively
|
Poole1, A.T., Murfet1, I.C. and Vaillancourt2, R.E. |
1Department of Plant Science 2CRC for Temperate Hardwood Forestry University of Tasmania, Hobart, Tasmania 7001, Australia |
Arumingtyas et al. (1) recently identified several new ramosus loci including rms-3 and rms-4. We present here evidence that rms-3 is in linkage group 1 and rms-4 in linkage group 7.
The data for rms-3 were obtained from crosses between line WL6042 (rms-3 a i) and Hobart lines 63 (Rms-3 A I) and 111 (Rms-3 A i). The data for rms-4 are from crosses between line K164 (rms-4 Pgd-pF Wa Aat-mF AldoF Gal-2S) and Hobart lines 6 (Rms-4 Pgd-pF wa Aat-mS AldoS Gal-2S) and 111 (Rms-4 Pgd-pS Wa Aat-mS AldoS Gal-2F). K164 is the type line for rms-4 and WL6042 is a representative line for rms-3 (1). The plants were grown in the glasshouse under an 18 h photoperiod following the procedures previously described (1). The joint segregation c2 values and recombination fractions were calculated using the LINKAGE-1 program (2). All crosses were fully fertile.
Rms-3 showed significant evidence of linkage with group 1 marker A in two crosses (Table 1). The combined data generate a linkage c2 = 21.17 (P = 0.000004) and a recombination fraction of 25 ± 8%. There was no evidence of linkage between Rms-3 and marker I in the lower section of group 1 (Table 1). The map position of the Rms-3 locus remains to be determined and we have made crosses involving several additional group 1 markers.
Table 1. Segregation data obtained from F2 populations for rms-3 and markers a and i in linkage group 1.
|
Genes |
Cross |
Phenotype |
Chi-square |
Recomb. Fract. |
SE |
||||||
|
DD |
DR |
RD |
RR |
Total |
Locus 1 |
Locus 2 |
Joint |
||||
|
Rms-3 A |
6042x111 |
39 |
5 |
12 |
8 |
64 |
1.33 |
0.75 |
6.97* |
30.9 |
11.1 |
|
|
6042x63 |
46 |
7 |
3 |
8 |
64 |
2.08 |
0.08 |
17.98** |
18.8 |
12.0 |
|
|
Combined |
85 |
12 |
15 |
16 |
128 |
0.04 |
0.67 |
21.17** |
25.0 |
8.2 |
|
Rms-3 I |
6042x63 |
37 |
16 |
8 |
3 |
64 |
2.08 |
0.75 |
0.04 |
|
|
|
A I |
6042x63 |
32 |
17 |
13 |
2 |
64 |
0.08 |
0.75 |
2.51 |
|
|
*,** P < 0.01 and 0.0001, respectively; all other cases P > 0.05.
Table 2. Segregation data obtained from F2 populations for rms-4 and several markers in linkage group 7.
|
Genes |
Cross |
Phenotype a |
Chi-square |
Recomb. Fract. |
SE |
||||||||||||
|
Locus 1 |
Locus 2 |
Joint |
|||||||||||||||
|
DD |
DR |
RD |
RR |
|
|
|
|
|
Total |
|
|
|
|||||
|
Rms-4 Wa |
164x6 |
39 |
10 |
14 |
1 |
|
|
|
|
|
64 |
0.08 |
2.08 |
1.52 |
34.0 |
10.9 |
|
|
|
|
DF |
DH |
DS |
RF |
RH |
RS |
|
|
|
|
|
|
|
|||
|
Rms-4 Pgd-p |
164x111 |
23 |
50 |
27 |
11 |
12 |
5 |
|
|
|
128 |
0.67 |
0.19 |
3.15 |
40.8 |
5.3 |
|
|
Rms-4 Aat-m |
164x111 |
3 |
26 |
22 |
11 |
0 |
0 |
|
|
|
62 |
1.74 |
3.68 |
45.85* |
5.8 |
3.0 |
|
|
|
164x6 |
1 |
31 |
16 |
14 |
1 |
0 |
|
|
|
63 |
0.05 |
0.05 |
52.51* |
3.3 |
2.3 |
|
|
|
Total |
4 |
57 |
38 |
25 |
1 |
0 |
|
|
|
125 |
1.18 |
1.94 |
98.10* |
4.4 |
1.9 |
|
|
Rms-4 Aldo |
164x111 |
7 |
43 |
21 |
11 |
8 |
1 |
|
|
|
91 |
0.44 |
1.68 |
21.15* |
21.0 |
4.7 |
|
|
|
164x6 |
9 |
21 |
16 |
11 |
4 |
0 |
|
|
|
61 |
0.01 |
2.51 |
16.19* |
21.7 |
5.9 |
|
|
|
Total |
16 |
64 |
37 |
22 |
12 |
1 |
|
|
|
152 |
0.32 |
0.00 |
37.23* |
21.3 |
3.7 |
|
|
Rms-4 Gal-2 |
164x111 |
25 |
54 |
15 |
4 |
16 |
7 |
|
|
|
121 |
0.47 |
3.79 |
2.37 |
40.0 |
5.4 |
|
|
Wa Aat-m |
164x6 |
14 |
26 |
13 |
1 |
6 |
3 |
|
|
|
63 |
2.80 |
0.05 |
1.25 |
42.9 |
7.6 |
|
|
Wa Aldo |
164x6 |
19 |
19 |
14 |
1 |
6 |
2 |
|
|
|
61 |
3.42 |
2.51 |
3.28 |
43.7 |
7.7 |
|
|
|
|
FF |
FH |
FS |
HF |
HH |
HS |
SF |
SH |
SS |
|
|
|
|
|
|
|
|
Pgd-p Aat-m |
164x111 |
4 |
7 |
3 |
9 |
9 |
8 |
4 |
11 |
7 |
62 |
1.06 |
3.68 |
2.37 |
45.5 |
6.3 |
|
|
Pgd-p Aldo |
164x111 |
6 |
5 |
7 |
15 |
26 |
10 |
4 |
13 |
7 |
93 |
0.29 |
1.65 |
5.23 |
47.6 |
5.2 |
|
|
Pgd-p Gal-2 |
164x111 |
4 |
15 |
10 |
21 |
39 |
15 |
8 |
7 |
7 |
126 |
0.14 |
5.35 |
6.34 |
42.9 |
4.3 |
|
|
Aat-m Aldo |
164x111 |
8 |
2 |
2 |
5 |
19 |
8 |
0 |
3 |
12 |
59 |
4.80 |
0.73 |
30.78* |
20.7 |
4.3 |
|
|
|
164x6 |
12 |
8 |
0 |
3 |
16 |
6 |
0 |
7 |
9 |
61 |
0.02 |
2.51 |
28.66* |
21.5 |
4.3 |
|
|
|
Total |
20 |
10 |
2 |
8 |
35 |
14 |
0 |
10 |
21 |
120 |
2.18 |
0.32 |
56.83* |
21.1 |
3.0 |
|
|
Aat-m Gal-2 |
164x111 |
1 |
4 |
7 |
8 |
16 |
10 |
4 |
6 |
4 |
60 |
3.20 |
1.20 |
4.07 |
39.8 |
6.1 |
|
|
Aldo Gal-2 |
164x111 |
1 |
12 |
8 |
10 |
37 |
11 |
7 |
13 |
2 |
101 |
5.42 |
2.25 |
9.14 |
32.8 |
4.2 |
|
a D = dominant, R = recessive, F = homozygous fast, H = heterozygous, and S = homozygous slow. The first named locus is shown first.
Numbers in bold face indicate the parental phenotypes.
* P <0.001; all other cases P >0.05.
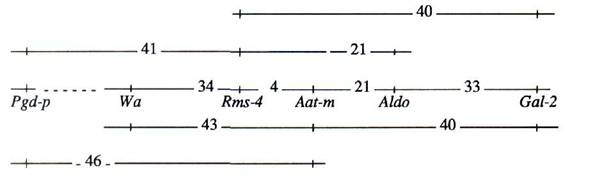
Rms-4 showed significant evidence of linkage with group 7 isozyme markers Aat-m and Aldo in two crosses (Table 2). The combined data indicate strong linkage between Rms-4 and Aat-m with a distance of about 4 cM. Unfortunately, the map position of Rms-4 relative to the other group 7 loci is not clear as the distances Rms-4 to Aldo and Aat-m to Aldo both work out at 21 cM and the distances Rms-4 to Gal-2 and Aat-m to Gal-2 both work out at 40 cM. The results for Pgd-p, Wa, Rms-4 and Aat-m indicate the order shown but these data are far too weak to permit a definite conclusion. The relative position of Rms-4 therefore remains to be determined.
The data from Table 2 may be plotted as follows:

This map is, in general, consistent with the latest pea map (3) but more recombination occurred between Wa and the Rms-4 and Aat-m loci than might be expected; neither joint segregation c2 is significant (P > 0.05). However, the sample size (n = 64 and 63) is fairly small for a morphological marker and larger numbers are necessary to properly establish these relationships.
In summary, Rms-3 is in linkage group 1 about 25 cM from A, and Rms-4 is in linkage group 7 about 4 cM from Aat-m, but the map position of both loci remains to be determined.
Acknowledgements. We thank Peter Bobbi, Katherine McPherson, Sarah G.A. Martin, Sarah K. Martin, Minh Nguyen and Scott Taylor for technical assistance and the Australian Research Council for financial support.
1.
Arumingtyas, E.X.,
Floyd, R.S., Gregory, M.J. and Murfet, I.C. 1992. Pisum
Genetics 24:17-31.
2. Suiter, K.A., Wendel, J.F. and Case, J.S. 1983. J. Hered. 74:203-204.
3.
Weeden, N.F., Swiecicki,
W.K., Ambrose, MJ. and Timmerman, G.M. 1993.
Pisum Genetics 25: Cover.