|
Pisum Genetics |
Volume 25 |
1993 |
Research Reports |
pages 27-31 |
Mapping of the new mutation blb and the problem of integrity of linkage group I
|
Kosterin, O.E. and Rozov, S.M. |
Institute of Cytology and Genetics Novosibirsk 630090, Russia |
A new recessive mutant, designated blb (bulbosus), is described and mapped on linkage group I.
The blb mutation was induced in line SGE following treatment with 0.15% EMS (ethylmethanesulphonate). The original mutant line was registered as SGE80. The initial line SGE was derived by V.A. Berdnikov from crosses among lines VIR6135 (from Greece), VIR320 (from Palestine, Pisum sativum syriacum), and an original ultra rapid line, Sprint-1.
The blb mutation transforms the shape of leaflets and stipules. The leaflets of homozygous blb plants are rather narrow, especially at lower nodes, and are wedge-shaped so that the maximal width of a leaflet is attained distally from the middle of its length (Fig. 1). At the first several nodes, the leaflet edges are rolled inside. The stipules are narrowed and, as a rule, do not overlap each other. The pods are narrowed, especially in the base, and seed productivity is reduced. In seedlings of the original mutant line SGE80 the stem is slightly thickened just above the first scale leaf. However, in the F2 progenies of three crosses involving this mutant (see below) this feature was much more pronounced. The seedlings homozygous for blb appeared to be considerably delayed in their growth, and their stems within the first 2-3 nodes were shortened and extremely inflated - up to 1 cm wide (Fig. 1). The name bulbosus was given to the mutant due to this unusual swelling of the stem. It was our impression that the apex ceased to grow and all the nutrients were directed into stem inflation. In the F2 progeny of cross SGE80 x WL1238, which was grown in a greenhouse, the apical buds of homozygous blb plants usually died and several lateral branches grew from basal nodes. In the F2 of cross VIR3971 x SGE80, which was planted in the field, mutant seedlings ceased growth for several days and then produced a normal shoot from the apical bud. In this cross the mutant plants usually produced no basal branches, while their counterparts with a normal phenotype exhibited vigorous basal branching. The situation was therefore opposite to that observed in the previous cross. The degree of stem inflation in mutant seedlings was variable and some blb homozygotes had stems only slightly thickened, as in the original mutant line SGE80. The modified leaflet shape was, however, an invariable character of blb homozygotes.
The F2 plants of cross SGE80 x WL1238 were tested for histone H1 electrophoretic spectrum, as described in (1, 2). The parental lines have different allelic variants of H1 subtype 7: SGE80 has allele His72 and WL1238 has allele His73. A significant linkage of 20 cM was detected between loci blb and His7 (Table 1) suggesting that blb belongs to linkage group I. Unfortunately, no other neighbouring marker genes were involved in this cross.
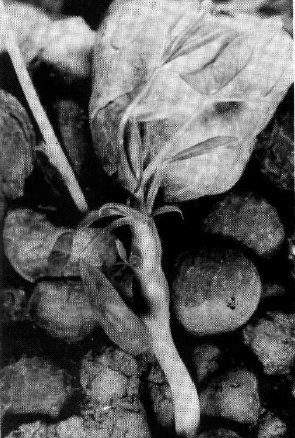
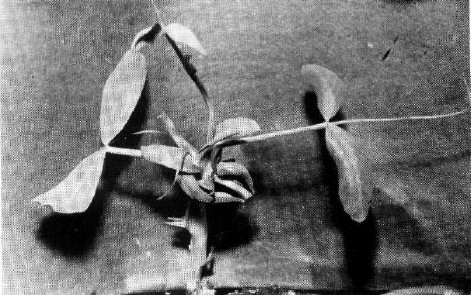
Figs la and b. Seedlings homozygous for blb.
Table 1. Joint segregation of genes His7 and blb in the F2 of cross SGE80 x WL1238.
|
|
His73 |
His73 / His72 |
His72 |
|
Blb |
15 |
37 |
7 |
|
blb |
1 |
5 |
11 |
Recomb. Fract = 19.91%; SE = 5.04%; Linkage c2 = 20.60; P = 0.00003.
In order to map gene blb, two other crosses were carried out: VIR3971 x SGE80 and SGE80 x OK7. Line SGE80 has alleles blb, His(2-6)1221 [this is the cluster of genes encoding histone H1 subtypes 2-6 (1, 2)], His72, A, Dw, and i; line VIR3971 has alleles Blb, His(2-6)1111, His71, A, Dco, and I; line OK7 (derived by O. Kosterin from Weibullsholm lines WL1393, WL1688, and WL102) has alleles Blb, His(2-6)1121, His73, a, Dco, and I. In both crosses, the F1 hybrids were vigorous and exhibited full fertility of both pollen and ovules. We grew F2 progenies comprising 663 plants of the former cross and 123 plants of the latter. F2 segregation data are presented in Tables 2 and 3.
The recombination frequencies inferred from the cross VIR3971 x SGE80 indicate the following arrangement of genes:
His(2-6)___________ His7 blb
____ 28.1 ± 1.5 17.6 ± 1.6
36.6 ± 2.2____________ cM
The distance between His(2-6) and His7 corresponds well to that obtained in several other crosses (2, 3, and unpub.).
The data from cross SGE80 x OK7 indicate the following map segment:
His(2-6) a His7 blb
13.6 ± 3.3 33.0 ± 4.0 18.7 ± 3.8