|
Pisum Genetics |
Volume 25 |
1993 |
Research Reports |
pages 18-21 |
A more precise location of the breakpoints in Hammarlund's K-line
|
Berdnikov, V.A., Gorel', F.L. and Temnykh S.V. |
Institute of Cytology and Genetics Novosibirsk 630090, Russia |
Hammarlund's K-line (L-84) is well known to contain a translocation between chromosomes 1 and 5 with the T-points being strongly linked to genes a, gp and cri (6). As first shown by Pellew (8) and then confirmed by others (6), a high frequency of tertiary trisomics occurs in the progeny of hybrids between the K-line and N-lines (normal structural type). Functional female (n+1) gametes as a rule contain chromosomes 1 and 5 from the N-parent plus the short interchange chromosome formed by the short arms of chromosomes involved (most probably chromosome 51). In this paper we have used the same numbers for chromosomes and for linkage groups to avoid confusion in designation.
We have used the phenomenon of tertiary trisomics to locate more precisely the breakpoints of the translocation in relation to adjacent markers on both the chromosomes involved. Of particular importance for our study was the highly polymorphic locus His (2-6) which encodes several subtypes of histone H1. This locus is tightly linked to gene a, and its alleles can easily be distinguished by gel electrophoresis (1). With appropriate combinations of different histone H1 alleles it is possible to recognise three doses of these genes (2).
Three testcrosses were performed by using F1 hybrids between the K-line and N-lines as females and tester N-lines as males. In our experiments we used WL-1476 from the Nordic Gene Bank collection which corresponds to Hammarlund's K-line. The standard line WL102 and our marker line 5-11, (both a, lf) were used as N-partners in testcross 1. A single F2 plant with genotype cri, gp from cross WL1238 x SGE182 (9), and inbred line 87-19 i-g originating from cross JI1794 x Slow (received from Dr N.F. Weeden), were involved in testcrosses 2 and 3. The phenotypes of progenies of the testcrosses are presented in Tables 1-3. Instead of the usual four digit designation for the His(2-6) phenotype, lower case letters are used to mark each different allelic combination of H1 bands. Trisomics (2N+51) were identified as plants having reduced pollen fertility (about 75% of normal pollen grains) and bracts (8).
The data in Table 1 show that the short interchange chromosome cannot contain genes a, lf and His(2-6) because the trisomics display the recessive phenotype for genes a and lf and they have only two allelic combinations of histone H1 (e.g. those carried by chromosome 1 of both N-lines, WL102 and 5-11). Two trisomic plants derived from testcross 2 demonstrate the same type of histone H1 pattern, namely they have a combination of two His(2-6) gene sets originating from parents with a normal karyotype. The phenotype of two plants arising from crossovers between a and lf (see Table 1) indicates that the position of the T-point is distal to either of these two markers (i.e. nearer to the telomere of the short arm of the chromosome).
Table 1. Phenotypes of progeny from testcross 1.
(WL1476 x WL102) x 5-11: His(2-6)a A Lf / His(2-6)b a lf a x His(2-6)c a lf a
|
Karyotype |
Phenotype |
Pollen fertility |
Number |
||
|
His(2-6) |
A |
Lf |
|||
|
2N |
His(2-6)b/His(2-6)c |
a |
lf a |
~100% |
18 |
|
2N |
His(2-6)b/His(2-6)c |
a |
Lf |
~100% |
2 |
|
N+K |
His(2-6)a/His(2-6)c |
A |
Lf |
~50% |
13 |
|
2N+51 |
His(2-6)b/His(2-6)c |
a |
lf a |
70-80% |
3 |
Table 2. Phenotypes of progeny from testcross 2.
(WL1476 x cri gp) x 87-19 i-g: His(2-6)a/His(2-6)d, Cri Gp/cri gp x His(2-6)e, Cri gp
|
Karyotype |
Phenotype |
Pollen fertility |
Number |
||
|
His(2-6) |
Cri |
Gp |
|||
|
2N |
His(2-6)d/His(2-6)e |
Cri |
gp |
~100% |
18 |
|
N+K |
His(2-6)a/His(2-6)e |
Cri |
Gp |
~50% |
14 |
|
2N+51 |
His(2-6)d/His(2-6)e |
Cri |
gp |
70-80% |
2 |
Table 3. Phenotypes of progeny from testcross 3.
(WL1476 x cri gp) x cri gp: His(2-6)a/His(2-6)d, Cri Gp/cri gp x His(2-6)d, cri gp
|
Karyotype |
Phenotype |
Pollen fertility |
Number |
|||
|
His(2-6) |
Cri |
Gp |
||||
|
2N |
His(2-6)d |
cri |
gp |
~100% |
7 |
|
|
N+K |
His(2-6)a/His(2-6)d |
Cri |
Gp |
~50% |
9 |
|
|
2N+51 |
His(2-6)d |
Cri |
gp |
70-80% |
2 |
|
As for the location of the breakpoint in relation to markers on chromosome 5, the data of the testcross 3 (Table 3) clearly indicate that the T-point lies between genes cri and gp, and that chromosome 51 contains gene Cri but does not carry Gp. Two trisomic plants derived from testcross 2 also have the recessive phenotype for gene gp. Progeny analysis of one trisomic plant originating from testcross 3 confirmed this result. Although the trisomics (2N+51), as a rule, are highly female sterile (8), one of our trisomic plants (No. 54) had a nearly normal level of fertility. We have harvested 41 seeds from that plant which gave rise to 31 cri and 10 Cri plants. Each Cri plant had bracts and reduced pollen fertility, and the additional chromosome registered as a univalent at metaphase 1 of the pollen mother cells (Fig. 1).
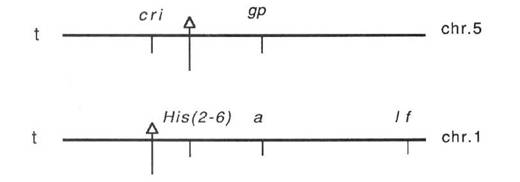
These data permit us to put the T-points (indicated by arrows) on the chromosome map as follows:

Here t stands for the telomeres of the short arms.
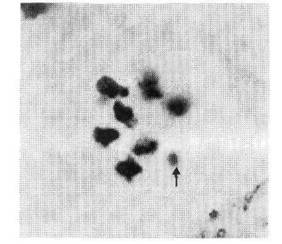
Fig. 1. Metaphase 1 in pollen mother cells of a tertiary trisomic 2N+51. Arrow shows the univalent 51.
Table 4. Percentage of crossingover between the T-points of Hammarlund's K-line and adjacent marker genes.
|
Cross |
Pair of factors |
Recomb. fract. ± SE |
Reference |
|
1+2 |
T - A |
4.2 ± 2.27 |
7 |
|
2 |
T - A |
7.7 ± 2.27 |
7 |
|
|
His(2-6) – A |
4.1 ± 1.5 |
1 |
|
A |
His(2-6) – A |
6.5 ± 1.1 |
3 |
|
B |
His(2-6) – A |
7.0 ± 2.2 |
3 |
|
|
His(2-6) – A |
5.3 ± 0.9 |
2 |
|
XS 38 |
T - Gp |
5.0 ± 3.29 |
10 |
|
XS 39 |
T - Cri |
1.0 ±0.82 |
10 |
|
|
T - Gp |
1.7 ± 0.29 |
4 |
We did not estimate the recombination frequency between the T-point and locus His{2-6) in our experiments directly but comparison of known data (Table 4) indicates that the distances of T to A and His(2-6) to A are approximately the same, and that the percentage of crossing-over varies in different crosses in the same range. The His(2-6) locus would be the nearest marker for the T-point of Hammarlund's K-line. The results of cytological studies of the K-line (6) suggest that the breakpoints are located close to centromeres, so this cluster of histone HI genes occupies the precentromeric region of chromosome 1.
Acknowledgements We thank Dr N.F. Weeden for providing seeds of the inbred line 87-19 i-j and useful discussion of our results.
Belyaev, A.I. and Berdnikov, V.A. 1981. Genetica (USSR) 17:498-504.
Gorel', F.L., Temnykh,
S.V., Lebedeva, I.P. and Berdnikov, V.A. 1992.
Pisum Genet. 24:48-51.
Kosterin, O.E. 1992. Pisum Genet. 24:56-59.
Lamm, R.L. 1974. PNL 6:29.
Lamm, R.L. 1976. PNL 8:36-37.
Lamm, R.L. 1986. PNL 18:34-37.
Lamm, R.L. and Miravalle, R.J. 1959. Hereditas 45:417-440.
Pellew, C. 1940. J. Genet. 39:363-390.
Rozov, S.M., Temnykh,
S.V., Gorel', F.L. and Berdnikov, V.A. 1993. Pisum
Genet. 25:46-51.
Snoad, B. 1966. Genetica Pol. 37:247-254.