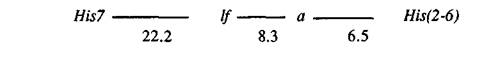
|
Pisum Genetics |
Volume 24 |
1992 |
Research Reports |
pages 56-59 |
Mapping of the third locus for histone H1 genes in pea
|
Kosterin, O.E.
|
Institute for Cytology and Genetics Novosibirsk 630090, Russia |
The electrophoretic pattern of histone H1 from inbred pea lines usually consists of seven bands, numbered according to their increasing electrophoretic mobility (Fig. 1). A screening of a world-wide collection of peas from the Vavilov All-Union Institute of Plant Breeding (VIR) revealed electrophoretic variants for each band. Each polymorphism exhibited a monogenic mode of inheritance and seven zones, each containing allelic polymorphism, were identified on polyacrylamide gels (2).
Two loci for histone H1 genes have been reported so far. A cluster of four tightly linked genes coding for bands in zones 3-6 was located on chromosome I, 4.1 ± 1.5 cM from a (1) (in that paper the preoccupied symbol H was proposed for the locus). The gene controlling variation in zone 2 was shown to be a member of the same gene cluster, and the entire cluster was given the gene symbol His(2-6) (4). A simple designation for allelic combination, or haplotype, of H1 bands 3-6 was adopted (2): a series of four digits corresponding to an allelic variant of zones 3, 4, 5, and 6, respectively. The allelic state of a gene coding band 2 is not included as the overwhelming majority of peas have the same variant of this band. If any zone lacks bands in a particular H1 spectrum, a zero is placed in the corresponding position in the haplotypic formula.
The gene producing variation in the most intense zone, zone 1, turned out to be 11 ± 3 cM from gene tl (4); symbol His1 was proposed for this gene. Unfortunately, in that paper the mutual location of genes r and tl in relation to other involved genes was stated erroneously (S.M. Rozov, personal communication). The location was corrected subsequently to 10.2 ±2.1 cM from gene r and 7.6 ±1.9 cM from gene tl (5).
The present communication concerns the mapping of the third histone H1 locus - that controlling variation in zone 7. This protein is strongly expressed only in actively growing apical parts of a seedling. Three electrophoretic variants in this zone were resolved (numbered 1, 2, and 3 according to their electrophoretic mobility). Tne majority of pea accessions possess variant 2. The frequencies of variants 1 and 3 in a sample of 272 accessions from VIR collection are 3.7% and 1.0%, respectively. In the progeny of crosses of plants with different variants they segregated as alleles of a single gene which is termed His7. The H1-containing protein fraction was isolated as in (4): about 300 mg of young leaves were homogenised in 200 ml of 0.15 M NaCl / 2 M urea / 0.1% Triton X-100 and the homogenate filtered and centrifuged at 1500 g for 5 min. The pellet was resuspended in 5 ml of 5% HClO4 and centrifuged again. To the supernatant was added 6 volumes of cold acetone / 0.5M H2SO4. The precipitated protein was centrifuged and dissolved in 0.9 M acetic acid / 8 M urea / and 15% (w/v) sucrose. Electrophoresis was carried out in 15% polyacrylamide / 0.5% N,N'-methylenbisacrylamide gel containing 6.25 M urea and 0.9 M acetic acid.
Linkage between His7 and His2-6 was documented in an F2 progeny (43 plants) from a cross between a line (obtained from the cross of cultivar Torsdag and entry VIR-4871, Georgia) with the 1323 haplotype of H1 zone 3-6 and allele His72 and tester line WL1238 (haplotype 1221 and His73). The recombination fraction was estimated by the method of maximum likelihood as being 15.5 ± 43%. The linkage was confirmed in the backcross (WL1393 x WL1688) x WL1393 (174 plants, recombination fraction 20.7 ± 3.0%).
In order to locate His7 more precisely, a double cross-over plant with haplotype 1323, variant His73 (Fig. la) and allele A of the anthocyanin locus was chosen from an F2 progeny of the cross WL1393 x WL1688. It was crossed with line WL102 having haplotype 1121, His72 (Fig. lc), a, and allele lfa determining (3) a low node of first flower (nodes 5-8, but mostly 6-7). The maternal plant produced the first flower at node 13, so it was assumed to possess allele Lf. All twelve F1 plants exhibited a hybrid H1 phenotype (Fig. 1b) corresponding to genotype 1323/1121, His72/His73 and had red flowers (A/a) which first appeared at the 13th-16th nodes. The F1 plants were backcrossed to WL102. The resulting progeny of 505 plants was analysed for the four segregating loci (A, Lf, His3-6, and His7). Two distinct classes were observed for node of first flower: genotype Lf/lfa flowered at nodes 13-17 and genotype lfa/lfa at nodes 5-8. In the latter class the lowest several flowers were often sterile.
The numbers of plants with 16 possible genotypes are given in Table 1 (Cross A). The recombination fractions calculated from these data (Table 2, Cross A) suggest the following arrangement of the genes:
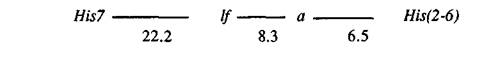
The configuration of genes lf, a and His(2-6) conflicts with that reported by Belyaev and Berdnikov (1), who placed the histone locus between lf and a. It should be noted that in their work no lines with identified lf alleles were used, and the allelic state of the gene was judged solely on the node of first flower.
In order to obtain additional information, another cross involving the same genes was analysed. Accession VIR-7094 (Peru), with haplotype 1221, alleles His73, A, and, possibly, Lfd (node of first flower 16-22) (3) was crossed with WL102 (1121, His72, a, lfa). Five F1 plants, exhibiting the expected hybrid phenotype, were back-crossed to WL102, producing a progeny of 142 plants. Segregation data for the four loci are given in Table 1 (Cross B). Similar genetic distances between the loci were obtained in this, second cross (Table 2, Cross B). Therefore I consider the gene order reported here as typical for garden pea. Belyaev and Berdnikov (1) may have been working with lines containing chromosomal rearrangements or the first flowering node might be determined in those crosses by interactions of lf with other segregating genes.
Thus, pea histone H1 is coded by a cluster of genes, His2-6, located near A on chromosome 1, and two additional genes. One of these additional genes, His7, is also located on chromosome 1, and its molecular product is most prominent in actively growing parts of a plant. The other gene, His1, is on a different chromosome, and its product comprises approximately half of pea histone H1 (Fig. 1.).
Table 1. Observed numbers of 16 possible genotypes in the progeny of two back-crosses involving genes A, Lf, His(2-6), and His7.
|
Cross A |
Cross B |
||||||||
|
Genes |
|
Genes |
|
||||||
|
Lf |
A |
His(2-6) |
His7 |
|
Lf |
A |
His(2-6) |
His7 |
|
|
Genotypes |
Numbers |
Genotypes |
Numbers |
||||||
|
lfa/lfa |
a/a |
1121/1121 |
2/2 |
179 |
lfa/lfa |
a/a |
1121/1121 |
2/2 |
51 |
|
lfa/lfa |
a/a |
1121/1121 |
2/3 |
46 |
lfa/lfa |
a/a |
1121/1121 |
2/3 |
15 |
|
lfa/lfa |
a/a |
1121/1323 |
2/2 |
12 |
lfa/lfa |
a/a |
1121/1221 |
2/2 |
4 |
|
lfa/lfa |
a/a |
1121/1323 |
2/3 |
8 |
lfa/lfa |
a/a |
1121/1221 |
2/3 |
1 |
|
lfa/lfa |
A/a |
1121/1121 |
2/2 |
1 |
lfa/lfa |
A/a |
1121/1121 |
2/2 |
0 |
|
lfa/lfa |
A/a |
1121/1121 |
2/3 |
0 |
lfa/lfa |
A/a |
1121/1121 |
2/3 |
0 |
|
lfa/lfa |
A/a |
1121/1323 |
2/2 |
14 |
lfa/lfa |
A/a |
1121/1221 |
2/2 |
4 |
|
lfa/lfa |
A/a |
1121/1323 |
2/3 |
4 |
lfa/lfa |
A/a |
1121/1221 |
2/3 |
1 |
|
Lf/lfa |
a/a |
1121/1121 |
2/2 |
5 |
Lfd/lfa |
a/a |
1121/1121 |
2/2 |
3 |
|
Lf/lfa |
a/a |
1121/1121 |
2/3 |
16 |
Lfd/lfa |
a/a |
1121/1121 |
2/3 |
5 |
|
Lf/lfa |
a/a |
1121/1323 |
2/2 |
0 |
Lfd/lfa |
a/a |
1121/1221 |
2/2 |
0 |
|
Lf/lfa |
a/a |
1121/1323 |
2/3 |
1 |
Lfd/lfa |
a/a |
1121/1221 |
2/3 |
1 |
|
Lf/lfa |
A/a |
1121/1121 |
2/2 |
4 |
Lfd/lfa |
A/a |
1121/1121 |
2/2 |
2 |
|
Lf/lfa |
A/a |
1121/1121 |
2/3 |
7 |
Lfd/lfa |
A/a |
1121/1121 |
2/3 |
2 |
|
Lf/lfa |
A/a |
1121/1323 |
2/2 |
44 |
Lfd/lfa |
A/a |
1121/1221 |
2/2 |
12 |
|
Lf/lfa |
A/a |
1121/1323 |
2/3 |
164 |
Lfd/lfa |
A/a |
1121/1221 |
2/3 |
41 |
|
Total |
|
|
|
505 |
Total |
|
|
|
142 |
Table 2. Recombination fractions between pairs of genes involved in the two crosses based on the data in Table 1.
|
Gene pairs |
Cross A |
Cross B |
|||
|
Recombination fraction |
Standard |
Recombination fraction |
Standard |
||
|
Lf |
A |
8.3 |
1.2 |
9.9 |
2.5 |
|
Lf |
His(2-6) |
14.1 |
1.6 |
15.5 |
3.0 |
|
Lf |
His7 |
22.2 |
1.9 |
23.9 |
3.6 |
|
A |
His(2-6) |
6.5 |
1.1 |
7.0 |
2.2 |
|
A |
His7 |
26.5 |
2.0 |
28.2 |
3.8 |
|
His(2-6) |
His7 |
27.5 |
2.0 |
29.6 |
3.8 |
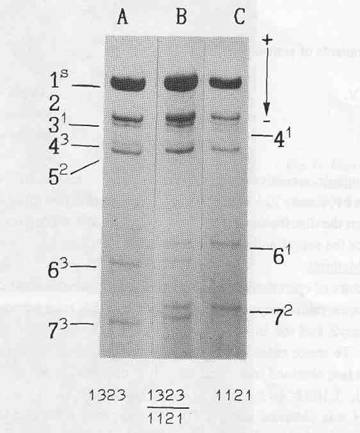
Fig. 1. Electrophoretic pattern of histone H1 isolated from: an individual plant chosen from the F2 progeny of the cross WL1393 x WL1688 - haplotype 1323, variant His73 (A); line WL102 - haplotype 1121, variant His72 (C); and their F1 hybrid (B). Figures denote subtypes, superscripts - their allelic variants.
Acknowledgements. I thank Drs S.M. Rozov and V.S. Bogdanova for providing preliminary information on the polymorphism for H1 zone 7 and Dr S. Blixt for providing seed of lines WL102, WL1238, WL1393, and WL1688 from the Nordic Gene Bank.
Belyaev, A.I., and Berdnikov, V.A. 1981. Genetika (USSR) 17:498-504.
Berdnikov, V.A., Bogdanova, V.S., Rozov, S.M. and Kosterin, O.E. 1989. In Vavilov's Heritage in Modern Biology. Nauka, Moscow, pp. 72-89. (in Russian).
Murfet, I.C. 1990. Pisum Newsl. 22:78-86.
Rozov, S.M., Bogdanova, V.S. and Berdnikov, V.A. 1986. Genetika (USSR) 22:2159-2166.
Smirnova, O.G., Rozov, S.M., Berdnikov, V.A. 1989. Pisum Newsl. 21:63-65.