STABLE TRANSFORMATION OF PEA
TISSUES AFTER CO-CULTIVATION WITH TWO AGROBACTERIUM TUMEFACIENS
STRAINS1
Filippone, E.
C.S. Miglioramento
Genetico Ortaggi
CNR, Portici, Italy
and P.F. Lurquin
Program in Genetics and Cell Biology
Washington State University, Pullman, USA
DNA transfer in plants is achieved
mainly by direct gene transfer using protoplasts or cells, or by
co-cultivation of plant tissues with Agrobacterium spp. harboring
Ti-plasmid-derived vectors. In both cases plant regeneration is necessary
to obtain transformed plants. As the genus Pisum is difficult to
regenerate in vitro, there is at present a lag in engineering this
genus. To overcome this problem, co-cultivation experiments were tried
using tissues showing the highest morphogenic ability, such as
meristematic tissues and epicotyl embryonic axes, on the basis of the
tissue culture technique and medium already set up by the authors
(2).
Seeds of pea cv. Puget were sown
in vitro in sterile conditions on PO medium (MS salts,
sucrose 10 g/l, agar 7 g/l, pH 5.8) and left to germinate at 24°C under 16
h light/8 h dark in a growth chamber. After three days, germinating
embryos were detached from the cotyledons and cut in three parts: apical
meristem, epicotyl segment and two cotyledonary node buds. These explants
were placed in contact with an overnight culture of Agrobacterium
tumefaciens for about two minutes and they were then transferred on P2
medium (MS salts, vitamins as Gamborg B5, sucrose 20 g/1, BAP 5 mg/l, IBA
1 mg/l, agar 7 g/l, pH 5.8). Two Agrobacterium strains were tested,
both carrying the pGA472 plasmid (1), containing the chimeric
NOS.NPTII.NOS gene which induces resistance to the neomycin antibiotic
group in transformed tissues. The 6044 strain was an A281-derived strain
that is hypervirulent on several solanaceous plants (3); the 6048 strain
is LBA4404, a non-oncogenic derivative of the wild-type strain Ach5,
harboring a Ti plasmid deleted of its T-region leaving the vir-region
intact (4). The co-cultivation stage was two-days long; after that,
explants were washed in P2 liquid medium containing claforan at 500 mg/1,
to stop the bacterial growth, and kanamycin at 100 mg/l, to start the
selection stage. Explants were subsequently dryed between two sterile
paper sheets and left to grow on P2 agarized medium with added antibiotics
as above. Explants were transferred monthly on fresh medium and, after 90
days of culture, transformation was scored on the basis of growing calli.
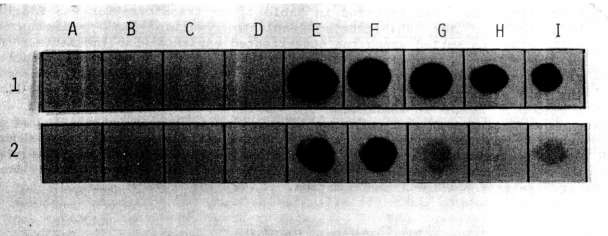
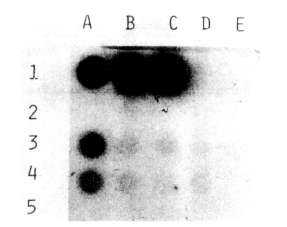
The presence of the NPTII gene in transformed tissues and its product were
checked, respectively, by dot blot and enzyme assay. The probe for the dot
blot was obtained by labeling the NPTII gene isolated from pABDI (kindly
supplied by J. Paszkowski) with dCT32P. As a positive control,
a line of Nicotiana tabacum transformed cells with the NPTII gene
was used.
As shown in Fig. 1 and Fig. 2, both
the dot blot and enzyme assays successfully demonstrated the stable
integration of the NPTII gene after