PNL Volume 19 1987 RESEARCH REPORTS
69
Sru, and Pur are mapped but the most useful variant may be the
isozymic locus Idh. Weeden and Marx (6) showed close linkage of
Idh with D and Pur with the suggestion that Idh is located between
these two. So a cross between Wt 11145 (Orc I orl D Idh-slow) x
Wt 11073 (orc i Orl d Idh-fast) was made at Wiatrowo. In F2
(field 1986) an undisturbed monohybrid segregation was found for
all genes (see Table 2A and the footnote). The electrophoretic
separation for Idh was done in the Biochemical Laboratory at
Wiatrowo.
isozymic locus Idh. Weeden and Marx (6) showed close linkage of
Idh with D and Pur with the suggestion that Idh is located between
these two. So a cross between Wt 11145 (Orc I orl D Idh-slow) x
Wt 11073 (orc i Orl d Idh-fast) was made at Wiatrowo. In F2
(field 1986) an undisturbed monohybrid segregation was found for
all genes (see Table 2A and the footnote). The electrophoretic
separation for Idh was done in the Biochemical Laboratory at
Wiatrowo.
The linkage of Orc with markers of chromosome 1 has been
confirmed, both with D and Idh (Table 2B). The CrO value for the
gene-pair Orc-D is almost identical with that from the previous
cross combination (Table IB). But despite undisturbed monohybrid
segregation there was no evidence of linkages for Orl in this
cross.
confirmed, both with D and Idh (Table 2B). The CrO value for the
gene-pair Orc-D is almost identical with that from the previous
cross combination (Table IB). But despite undisturbed monohybrid
segregation there was no evidence of linkages for Orl in this
cross.
If Idh is located between D and Pur (6), then the data from
Tables IB and 2B suggest the following gene order: Pur - Idh - D -
Orc - Orl?
Tables IB and 2B suggest the following gene order: Pur - Idh - D -
Orc - Orl?
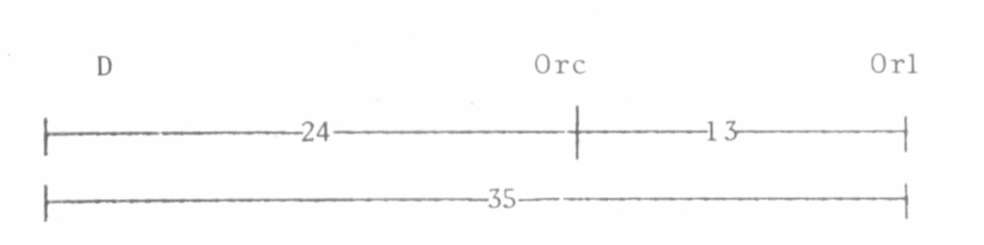
Fig. 1. The map distances in D segment on chromosome 1
Acknowledgement:
I wish to express my sincere gratitude to Mr. B. Wolko, M. Sc.
from Wiatrowo's Biochemical Laboratory, for electrophoretic sepa-
ration of Idh.
from Wiatrowo's Biochemical Laboratory, for electrophoretic sepa-
ration of Idh.
1. Blixt, S. and W. K. Swiecicki. 1983. PNL 15:9-10.
2. Ludwicki, J. and W. K. Swiecicki. 1983. PNL 15:41-42.
3. Ludwicki, J. and W. K. Swiecicki. 1985. PNL 17:51-53.
4. Swiecicki, W. K. 1982. Documentation of Genetic Resources:
A Model. S. Blixt, J. T. Williams, eds., NGB/IBPGR, Rome,
pp. 59-64.
A Model. S. Blixt, J. T. Williams, eds., NGB/IBPGR, Rome,
pp. 59-64.
5. Swiecicki, W. K. and S. Blixt. 1984. PNL 16:70-72.
6. Weeden, N. F. and G. A. Marx. 1984. J. Heredity 75:365-370.
xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx
x Editor's Note: x
x Editor's Note: x
x Recently, while inspecting seed samples of about a dozen x
x USDA Plant Introduction (P.I.) accessions, I noticed that x
x P.I. 143484 (from Iran) has orange cotyledons. x
x USDA Plant Introduction (P.I.) accessions, I noticed that x
x P.I. 143484 (from Iran) has orange cotyledons. x
xxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxxx
*****