76
PNL Volume 17
1985
RESEARCH REPORTS
|
|
||||
|
PNL Volume 17 1985
|
RF,SEARCH REPORTS
|
75
|
||
|
|
||||
|
RELATIVE POSITION OF Adh-1 AND Gal-3 ON CHROMOSOME 3
Weeden, N. F., G. A. Marx, NYS Agricultural Experiment Station
Geneva, NY USA
and E. Pagowska Experiment Station of Plant Breeding
and Acclimatization, Przebedowo, Poland
Previous mapping studies have identified three Loci which code (or
specific enzymes (Lap-1, Lap-2, and Acp-3) on chromosome 1 (1, 2).
These loci span most of the known linkage map ol this chromosome and
should be useful markers for further mapping studies. In this communi-
cation we report the position of two additional enzyme loci, Adh-1 and
Gal-3, relative to those already mapped and to the two morphological
markers St and B.
Starch gel electrophoresis was performed on seed, young leaf, and
flooded root tissue as described previously (2, 3, 4). Seed extracts
were primarily used to obtain acid phosphatase (AcP), beta-galactosidase
(Gal) and leucine aminopeptidase (Lap) phenotypes. Alcohol
dehydrogenase phenotypes were determined using extracts from root tissue
which had been submerged in water for 12 hours. Young leaf tissue was
used to confirm the Acp, Gal and Lap phenotypes. Only small amounts of
tissue were required for each sampling so that a single plant could be
assayed for all isozyme systems and still be grown to maturity in the
greenhouse.
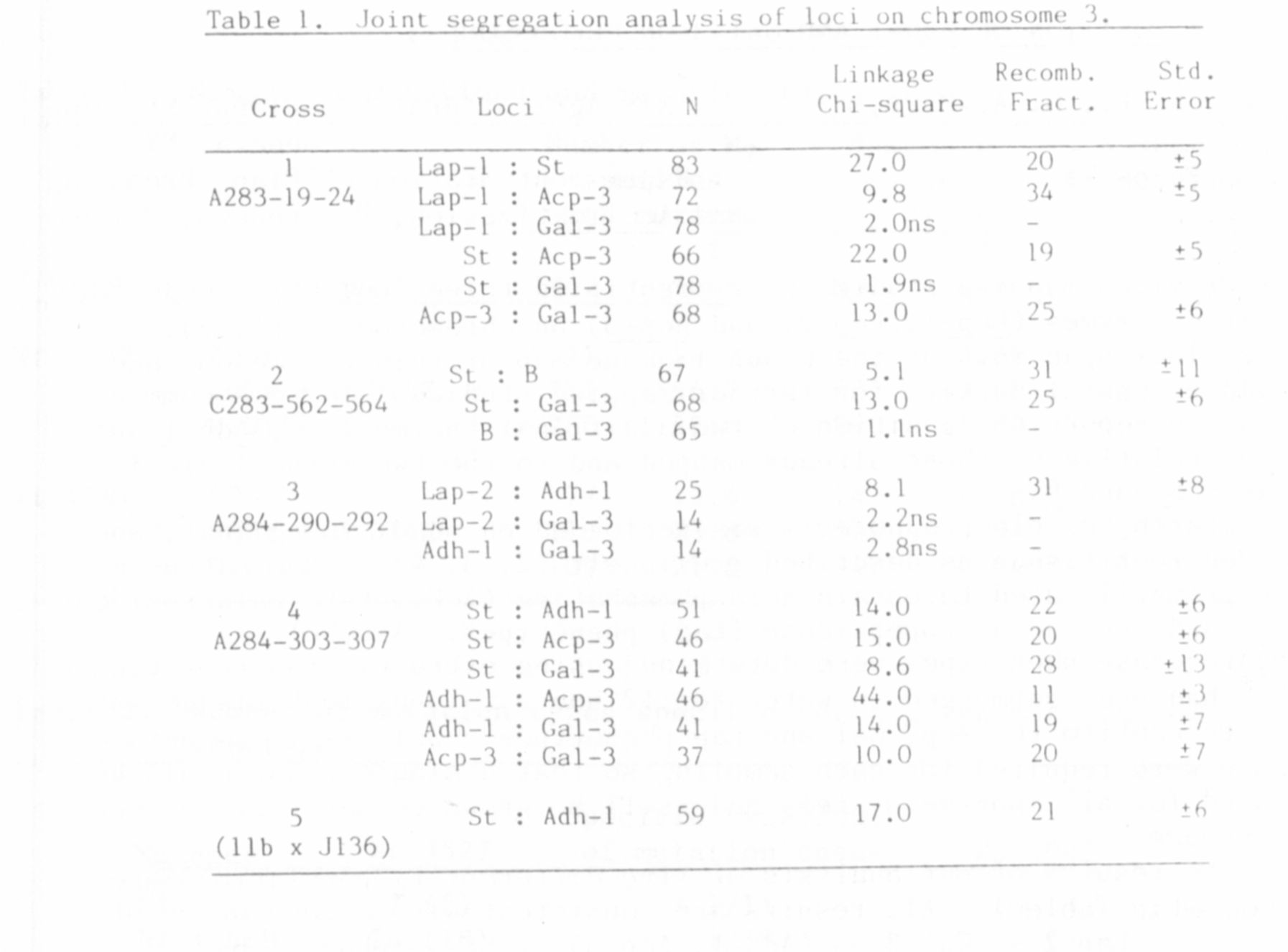
The results of our analysis of five different F2 populations are
presented in Table 1. All results are consistent with a gene order of
Lap 2 — Gal-3 — (Adh-1 , Acp-3) -- St -- (Lap-1, B).
Despite the significant recombination frequency observed between Adh-1
and Acp-3 in crosses #3 and #4, we were not able to determine which of
these loci was closer to St. We therefore place the two 1oci together
within parentheses to indicate that their relative sequence is unknown.
The loci Lap-1 and B were treated similarly although previous work
suggests that Lap-1 should be on the St side of B (2).
The map distances calculated between loci were relatively
reproducible when results from different crosses were compared. The St,
— (Adh-1, Acp-3) was calculated as 10, 20, and 10 recombinant units in
crosses 1, 4, and 5, respectively. Similar consistency was observed for
the St — Gal-3 and Gal-3 — (Adh-1, Acp-3) map distances. Ihe greatest
source of error probably was in the scoring of the Acp 1 phenotypes, for
these are faint in leaf tissue extracts and relatively poorly resolved
in seed extracts. Thus Adh-1 would appeal to be a better genetic marker
than Acp-3 for this particular region of chromosome 3.
|
||||
|
|
||||
|
1. Almgard, G. and K. Ohlund. 1970. PNL 2:9.
2. Weeden, N. F. and G. A. Marx. 1984. I. Hered. 75: 365-370.
3. Weeden, N. F. 1985. PNL 17:76-78.
4. Weeden, N. F. and E. Pagowska. 1985. PNL 17:79+80.
|
||||
|
|
||||
|
|
||||||
|
76
|
PNL Volume 17
|
1985
|
RESEARCH REPORTS
|
|||
|
|
||||||
 |
||||||
|
|
||||||
|
|
||||||