PNL Volume 17 1985 RESEARCH REPORTS 61
COMPARISON OF PROTEIN AND ENZYME PATTERNS IN SEEDS FROM FIELD- AND
PHYTOTRON-CULTIVATED PLANTS.
Muller, H. P. Institute of Genetics, University of Bonn
Federal Republic of Germany
Use of controlled environment facilities allows study of the
genetic control of the photoperiodic and thermoperiodic reactions of
various genotypes. Mature seeds from numerous genotypes with differing
growth and flowering behavior were analyzed electrophoretically with
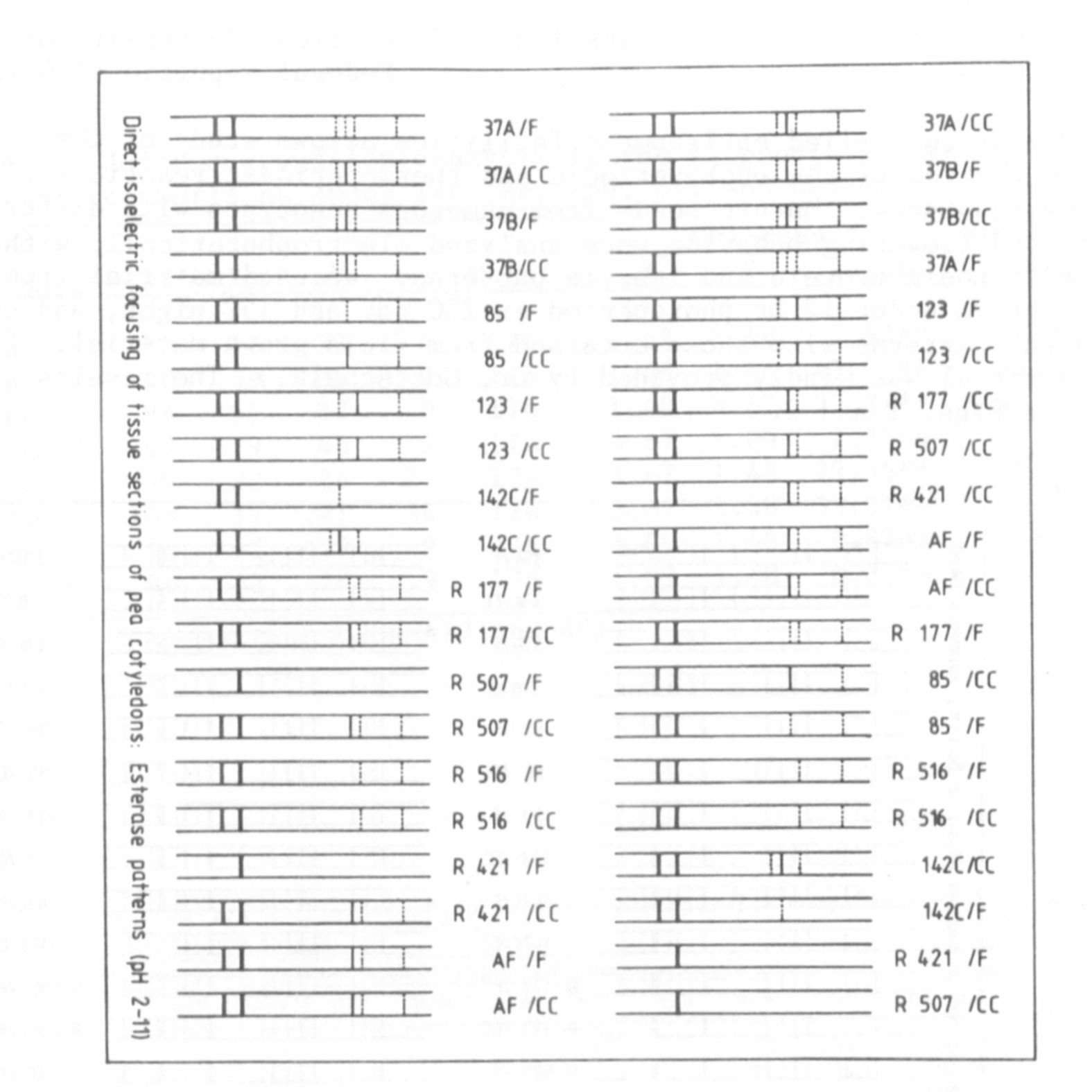
regard to their protein and isozyme patterns. We used material from
plants grown under 12 hr photoperiod at 25C day and 15C night, and com-
pared the patterns with those obtained from field grown material. (The
seed material was kindly provided by Dr. Gottschalk.) The results are
shown in Figs. 1 and 2.
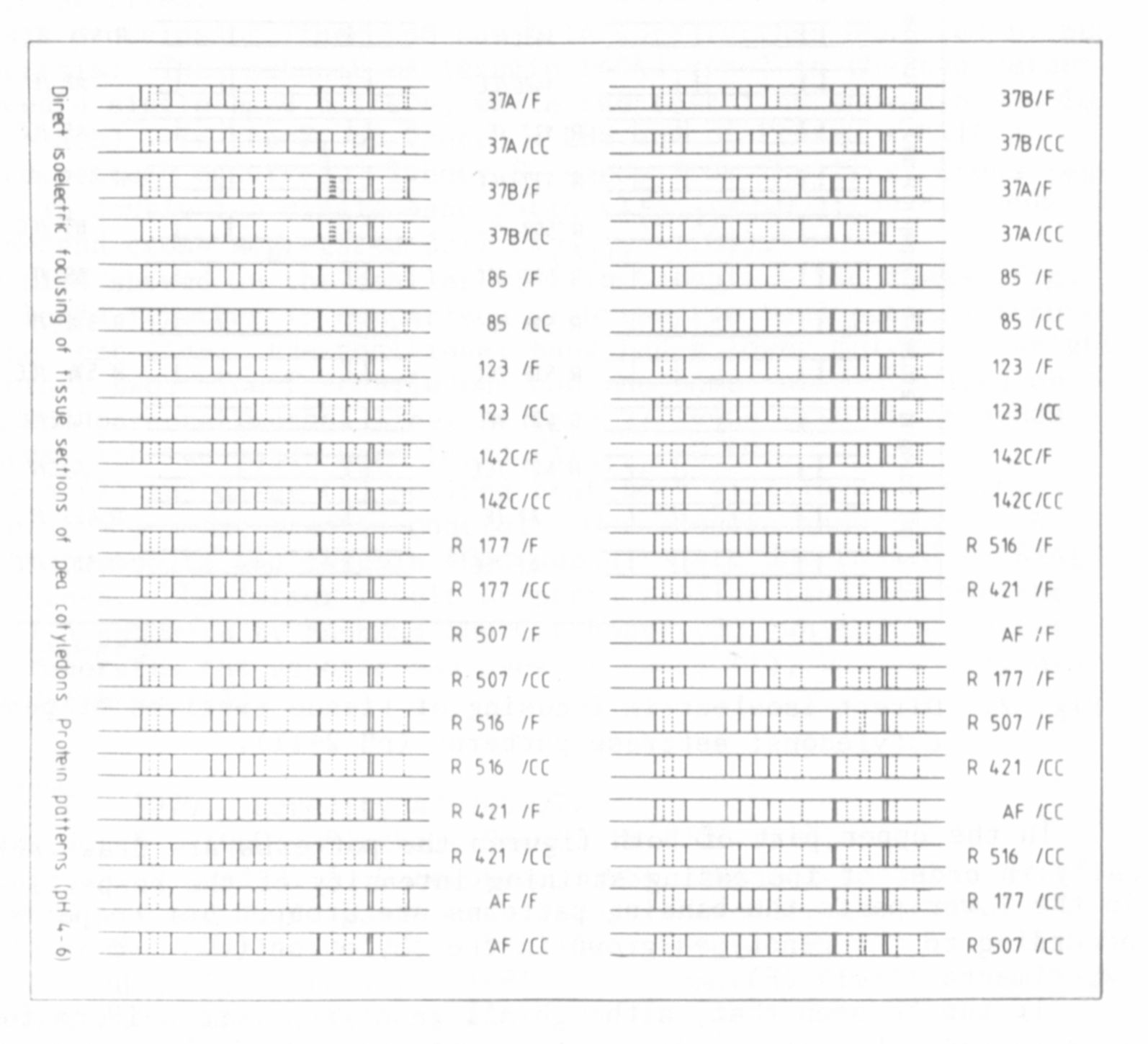
Fig. 1. Direct isoelectric focusing of tissue sections of pea
cotyledons: protein patterns (pH gradient 4-6).