PNL Volume 1M 1982 RESEARCH REPORTS 47
INFLUENCE OF NON-ALLELIC INTERACTIONS ON THE PHENOTYPE OF na
Marx, G. A. NYS Agricultural Experiment Station, Geneva, NY USA
A preliminary experiment was conducted to study the linkage rela-
tions and interactions of nana (na) and microdwarf (lm), two genes known
to reside in chromosome 6 (1,5,6). The na. parent derived from the na
mutant isolated at Geneva (2); it also carried other chromosome 6
markers, viz. Arg. Pl, and wlo. WL 1329, obtained from Dr. Blixt, was
used as a source of lm. In Blixt's inventory WL 1329, besiges being
identified as lm, is listed in the "genotype" category as crys , but in
the "origin" category as "microcryptodwarf". Unlike the na parent,
WL 1329 is arg pl and Wlo. Thus the known or presumed genotypes of the
two parents were:

The populations also segregated and were scored for genes which were
not relevant to the study, and, except to say that the genes segregated
according to expectation, they will not be considered further. Each of
two F2 populations examined descended from an individual plant grown
in the field. The two parents, additional F1 plants, and F2 populations
then were planted at the same time and grown in the same glasshouse
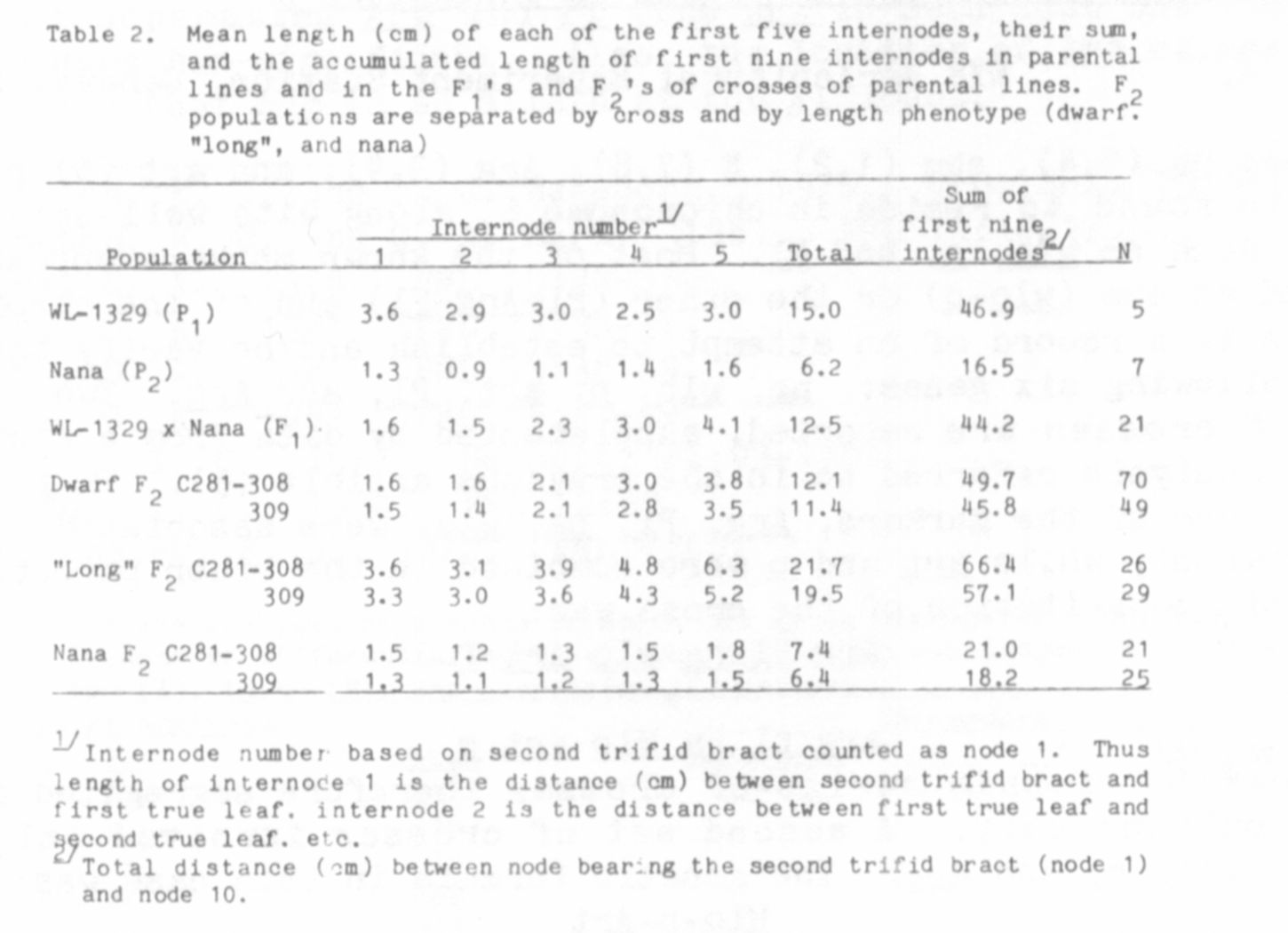
where they were fully classified and measured for internode length.
Nodes to flower were also recorded. A four-point linkage analysis of
Arg-Pl-Wlo-Na was performed on the combined populations and is presented
in the following article. Arg or Pl are not immediately relevant to
this article.
All F1's were classified as dwarf. The individual and combined F2
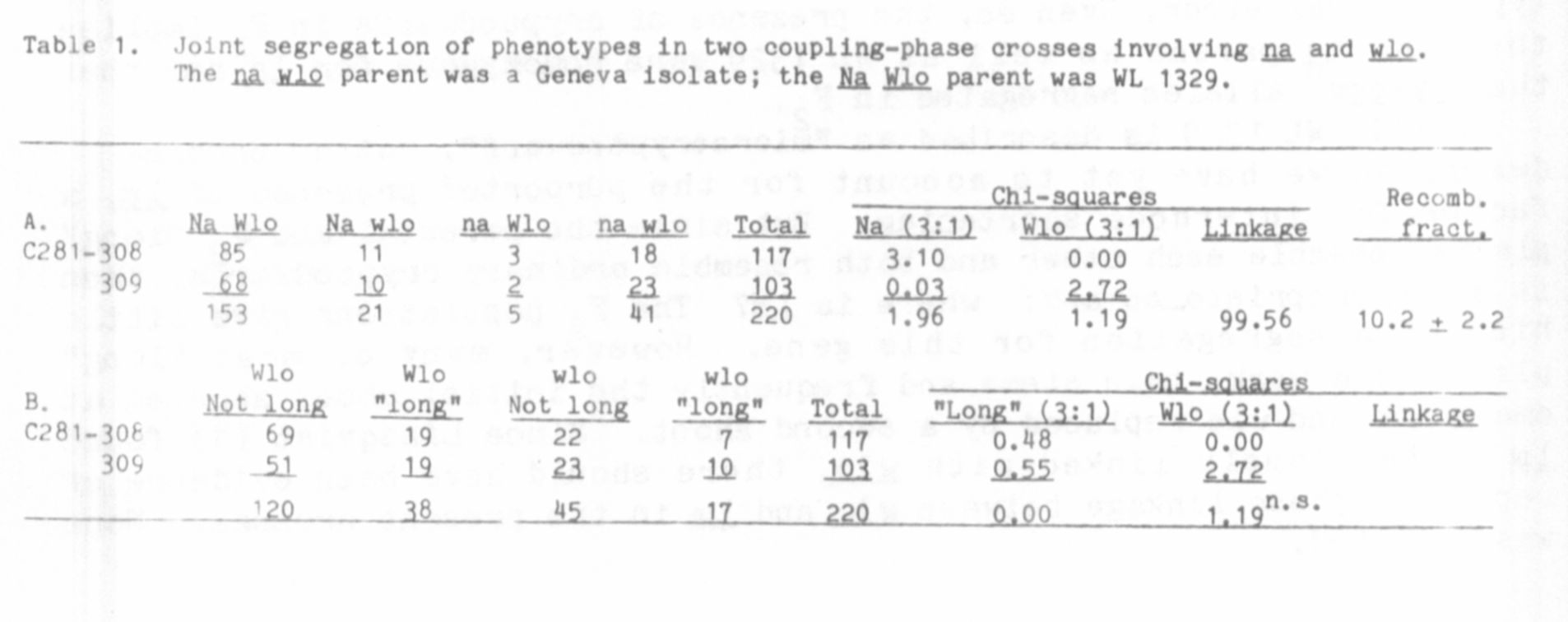
distributions (Table 1) reaffirm the known linkage between na. and wlo.
but the estimate of linkage intensity (10.8 ±_ 2.2) was somewhat higher
than other estimates (2,3). The distributions do, in fact, appear dis-
turbed, the Na wlo class being noticeably larger than the na Wlo class.
Despite this, segregations for either gene in the individual and com-
bined F2 populations were statistically not significantly different from
expected.